Fig. 1
- ID
- ZDB-FIG-170921-64
- Publication
- Uriu et al., 2017 - A framework for quantification and physical modeling of cell mixing applied to oscillator synchronization in vertebrate somitogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
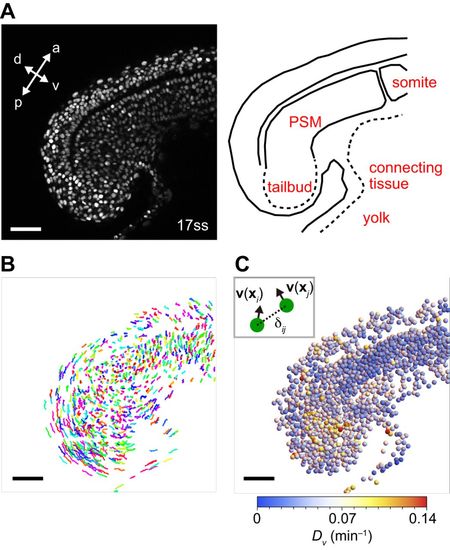
Quantification of cell mixing by the average directional derivative modulus of cell velocity vectors. (A) Left: Snapshot of a 17 ss embryo, with nuclei labeled with Histone h2AflV-gfp. a, anterior; p, posterior; d, dorsal; v, ventral. Right: schematic picture of PSM and tailbud. See also Movie 1. (B) Cell trajectories for the embryo shown in A, obtained by nuclear detection and tracking algorithms. Trajectories from time frame 1 to 20 (16.7 min) plotted in three-dimensional space. Each trajectory is assigned a color randomly. (C) Spatial profile of average directional derivative modulus of cell velocity vectors Dv for the 17 ss embryo shown in A. Spheres represent the positions of cells (radius chosen for better visibility). Scale bars: 50 μm. |