- Title
-
Normal locomotion in zebrafish lacking the sodium channel NaV1.4 suggests that the need for muscle action potentials is not universal
- Authors
- Akiyama, C., Sakata, S., Ono, F.
- Source
- Full text @ PLoS Biol.
|
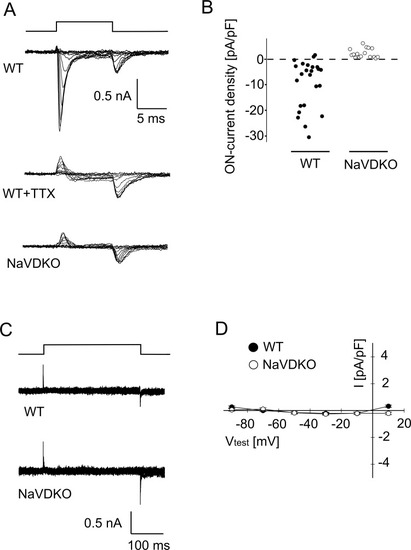
Electrophysiology of isolated myocytes. PHENOTYPE:
|
|
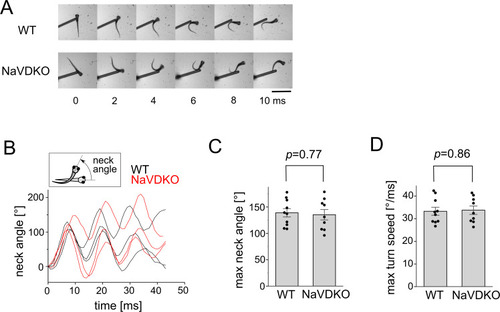
Escape response of WT and NaVDKO zebrafish. PHENOTYPE:
|
|
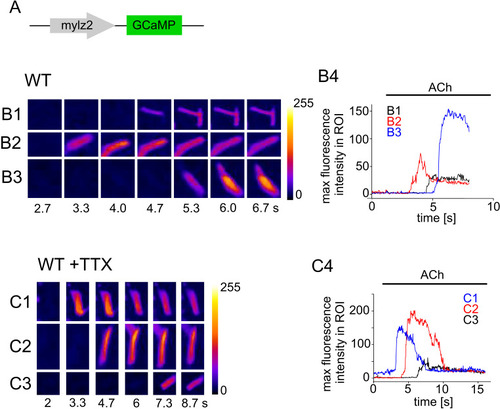
Ca2+ imaging of isolated myocytes in the absence and presence of TTX. |
|
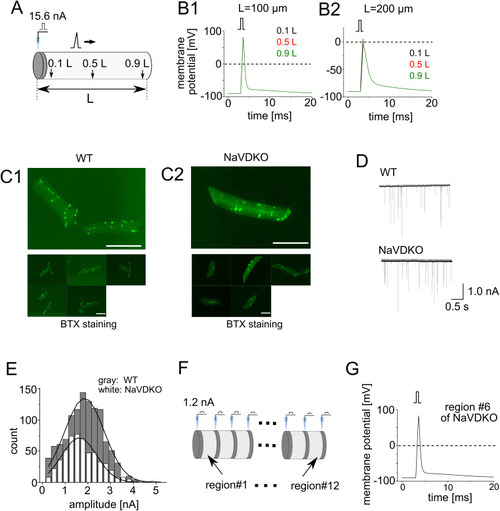
Mathematical simulation of depolarization in embryonic myocytes. |