- Title
-
ctdsp2 Knockout Induces Zebrafish Craniofacial Dysplasia via p53 Signaling Activation
- Authors
- Xia, X., Song, W., Zhang, F., Fan, Y., Zhang, B., Chen, X.
- Source
- Full text @ Int. J. Mol. Sci.
|
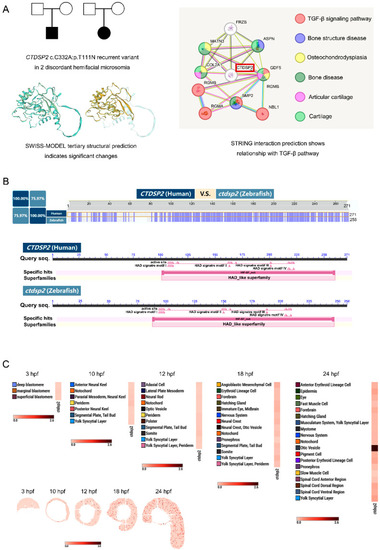
Identification of |
|
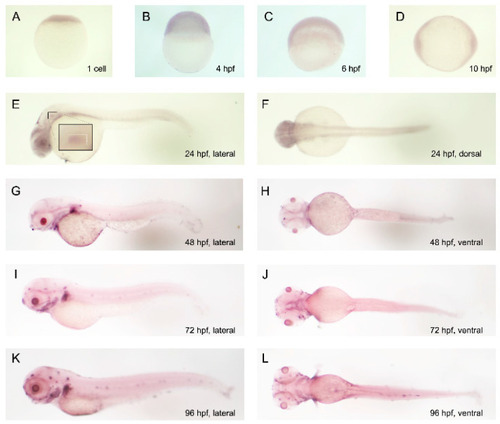
Temporal and spatial expression patterns of |
|
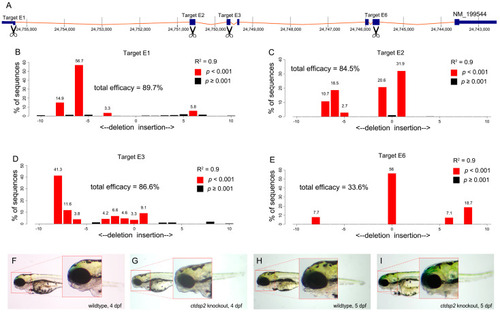
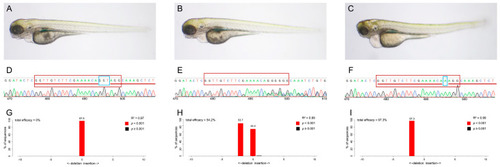
Knockout of |
|
( |
|
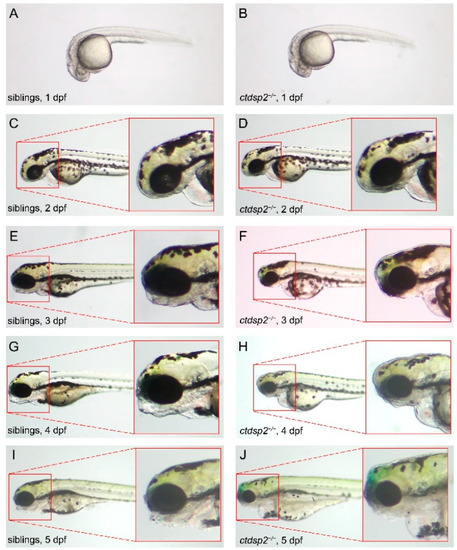
Phenotypic comparison between |
|
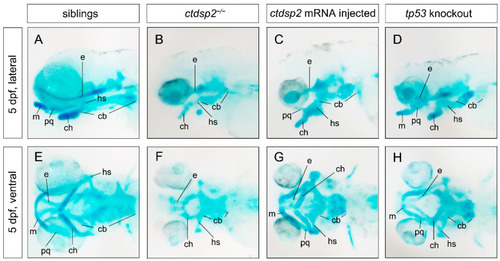
Staining results of pharyngeal arch cartilages using Alcian blue in various groups: ( |
|
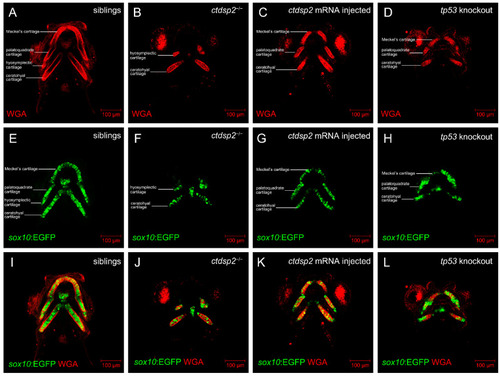
Comparative analysis of chondrocyte morphology in ( |
|
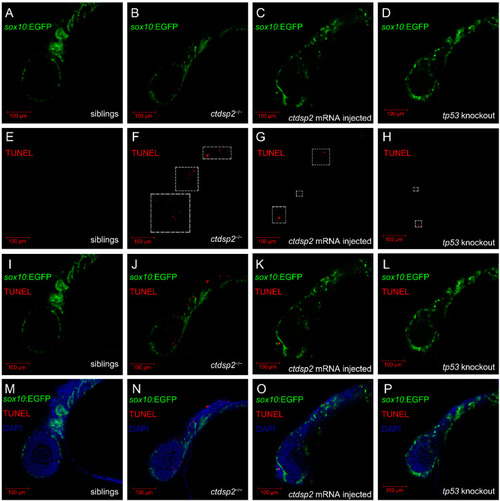
Analysis of somatic cell apoptosis in ( |
|
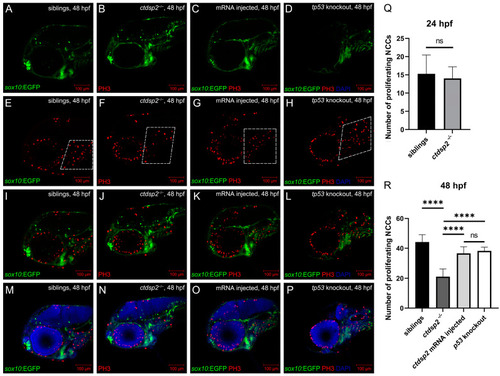
Immunofluorescence results depicting NCC proliferation through antiphosphohistone H3 (PH3) staining in ( |
|
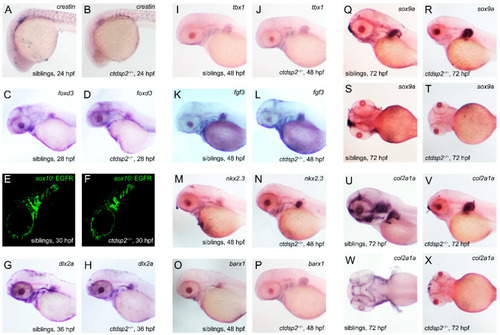
The impact of |
|
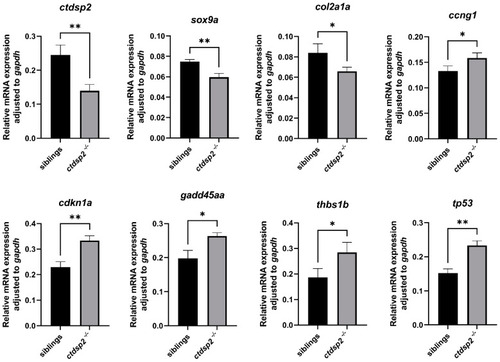
qPCR analysis of gene expression in |