- Title
-
Cross-species comparison reveals that Hmga1 reduces H3K27me3 levels to promote cardiomyocyte proliferation and cardiac regeneration
- Authors
- Bouwman, M., de Bakker, D.E.M., Honkoop, H., Giovou, A.E., Versteeg, D., Boender, A.R., Nguyen, P.D., Slotboom, M., Colquhoun, D., Vigil-Garcia, M., Kooijman, L., Janssen, R., Hooijkaas, I.B., Günthel, M., Visser, K.J., Klerk, M., Zentilin, L., Giacca, M., Kaslin, J., Boink, G.J.J., van Rooij, E., Christoffels, V.M., Bakkers, J.
- Source
- Full text @ Nat Cardiovasc Res
|
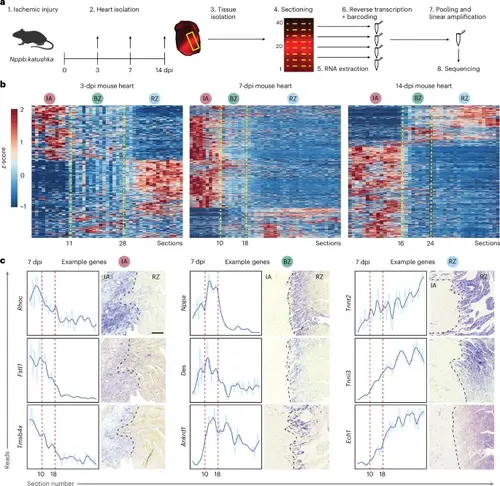
TOMO-seq reveals transcriptionally distinct regions in the injured mouse heart. a, Schematic overview of TOMO-seq workflow on injured mouse hearts. b, Three-dpi, 7-dpi and 14-dpi heatmaps showing hierarchical clustering for genes with a clear expression peak (z-score > 1 in more than four consecutive sections). Genes are on the y axis, and section numbers are on the x axis. Each section represents 100 μm of tissue. IA, BZ and RZ indicate consecutive sections with distinct gene profiles, separated by yellow dotted lines. c, Seven-dpi TOMO-seq plots with paired ISH images showing three representative genes for each zone. A total of n = 3 hearts were analyzed per staining. Red dashed lines in TOMO-seq plots indicate borders between IA, BZ and RZ, and black dashed lines in images indicate the border of the IA. Scale bar, 200 μm, which is the same for all ISH images. |
|
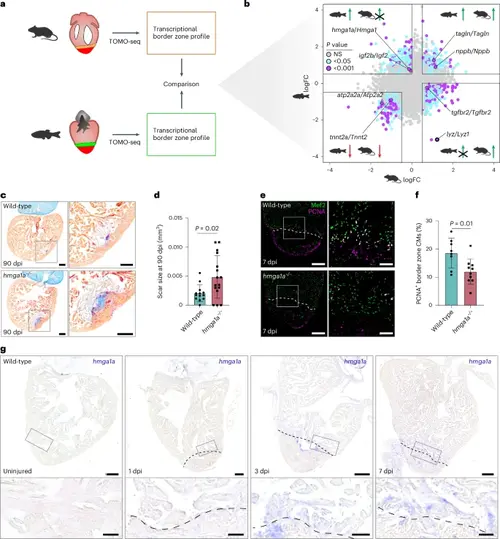
Interspecies comparison identifies Hmga1a, which spatially and temporally correlates with cardiac regenerative capacity. a, Schematic overview of the spatially resolved transcriptomic comparison of injured zebrafish and mouse BZs. b, Scatterplot analysis comparing BZ expression as logFC for homologous gene pairs. Gene pairs were selected based on the following criteria: only up in zebrafish (upper left quadrant); zebrafish logFC > 0.5, P < 0.05, and mouse logFC < 0; up in zebrafish and mouse (upper right quadrant); zebrafish logFC > 0.5, P < 0.05, and mouse logFC > 0.5, P < 0.05; down in zebrafish and mouse (lower left quadrant); zebrafish logFC < −0.5, P < 0.05, and mouse logFC < −0.5, P < 0.05; and only up in mouse (lower right quadrant); zebrafish logFC < 0 and mouse: logFC > 0.5, P < 0.05. Statistics were obtained using the R package edgeR, which uses GLMs and empirical Bayes methods to identify differentially expressed genes. NS, not significant. c, Representative images of AFOG staining on 90-dpi wild-type and hmga1a−/− zebrafish hearts, showing muscle in orange, fibrin in red and collagen in blue. Scale bars, 100 μm. d, Quantification of scar size in wild-type (n = 13) and hmga1a−/− (n = 16) hearts at 90 dpi. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test (P = 0.02). e, Representative images of immunofluorescent staining against Mef2 and PCNA on 7-dpi wild-type and hmga1a−/− zebrafish hearts. Dashed line indicates border with the injury. Overview scale bars, 100 μm; zoom-in scale bars, 20 μm. f, Quantification of proliferating BZ CMs in wild-type (n = 8) and hmga1a−/− (n = 10) hearts at 7 dpi. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test (P = 0.01). g, Representative images of ISH against hmga1a in uninjured, 1-dpi, 3-dpi and 7-dpi zebrafish hearts. n = 3 hearts were analyzed per condition. Scale bars, 100 μm in overviews and 25 μm in zoom-ins. Dashed line indicates border with the injury. |
|
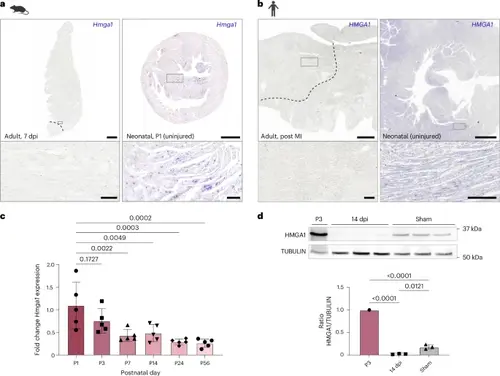
Hmga1 correlates with regenerative capacity of the mammalian heart. a, Representative ISH for Hmga1 in left ventricular tissue of injured adult mouse hearts (left panel) and in uninjured neonatal P1 mouse hearts (right panel). n = 3 hearts were analyzed per condition. Scale bars, 0.5 mm in the overview and 50 μm in the zoom-ins. Dashed line in the left panel indicates the injury border. b, Representative ISH images of HMGA1 expression in intraventricular septum tissue of an injured adult human heart (left panel) and in an uninjured neonatal human heart (right panel). n = 1 heart was analyzed per condition. Scale bars, 3 mm in the overview and 250 μm in the zoom-ins. Dashed line in the left panel indicates the infarct border. c, qPCR results for Hmga1 on cDNA libraries from whole mouse hearts at different postnatal timepoints. GAPDH was used as a reference gene. Five biological replicates were used per timepoint. Datapoints represent individual biological replicates. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Dunnett’s multiple comparison test. One-way ANOVA analysis indicates a significant difference in Hmga1 expression between different timepoints (P = 0.0002). Dunnett’s multiple comparison test shows that P7 (P = 0.0022), P14 (P = 0.0049), P24 (P = 0.0003) and P56 (P = 0.002) significantly differ from the P1 timepoint, whereas P3 does not (P = 0.1727). d, Western blot for HMGA1 on protein lysate from 14-dpi ventricles (n = 3) and sham ventricles (n = 3) compared to a P3 ventricle (n = 1). TUBULIN was used as a control protein. Ratios were calculated using TUBULIN. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show that both 14-dpi samples (P < 0.0001) and sham samples (P < 0.001) significantly differ from the P3 sample. Fourteen-dpi samples and sham samples do not significantly differ from each other (P = 0.021). |
|
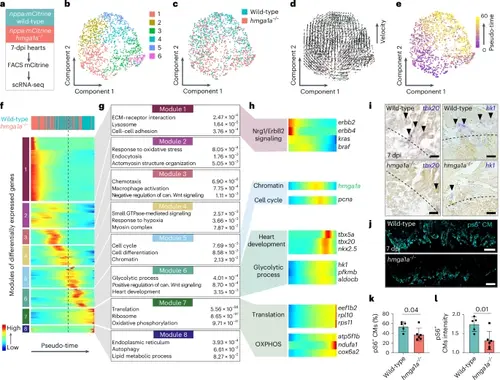
Hmga1a regulates progression of the regeneration program in BZ CMs. a, scRNA-seq workflow on nppa:mCitrine+ cells from 7-dpi wild-type (n = 12) and hmga1a−/− (n = 12) hearts. b, UMAP representation of scRNA-seq data after unsupervised clustering by Seurat. c, Distribution of wild-type and hmga1a−/− cells over UMAP. d, RNA velocity plotted on UMAP. Arrows are vectors that indicate the present and future position of a cell in the UMAP based on the ratio of spliced and unspliced reads. e, Pseudo-temporal ordering of cells performed using Monocle 2 represented on UMAP. Scale from 0 to 60 (purple-yellow). f, Self-organizing heatmap of gene modules co-expressed over pseudo-time. Each line represents a single gene. Cells ordered based on pseudo-time are divided in 100 bins. Legend on the top shows the predominant genotype present in each bin. Dashed line indicates the beginning of module 5, containing hmga1a. g, Three representative GO terms and their P value (obtained using DAVID online GO analysis tool) are shown for genes in modules 1–8 of f. can., canonical. h, Heatmaps over pseudo-time of individual genes of interest from modules 1, 5, 6 and 7. Linked biological processes are indicated on the left of the heatmaps; gene names are indicated on the right. i, Representative images from ISH against tbx20 (left) and hk1 (right) showing part of the BZ of wild-type and hmga1a−/− hearts at 7 dpi. n = 3 hearts were analyzed per condition. Dashed line indicates the injury border. Arrowheads indicate tbx20/hk1-expressing CMs. Scale bars, 50 μm. j, Representative images from immunofluorescent staining showing myocardial pS6 in the BZ of wild-type and hmga1a−/− zebrafish hearts at 7 dpi. Scale bars, 100 μm. k,l, Quantification of myocardial pS6 signal in wild-type (n = 5) versus hmga1a−/− (n = 6) 7-dpi BZ. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test and show a significant difference in percentage of pS6+ area relative to tropomyosin+ area (k) (P = 0.04) and in intensity of pS6 signal relative to tropomyosin signal (l) (P = 0.01). |
|
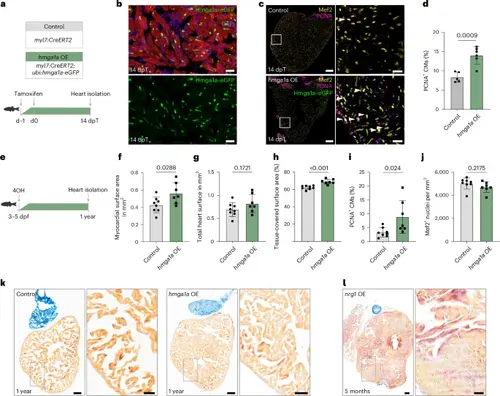
hmga1a OE stimulates CM proliferation resulting in myocardial expansion without pathological remodeling. a, Workflow of tamoxifen treatment for 14 days of hmga1a OE used in b–d. b, Representative images of immunofluorescent staining for GFP, α-actinin and DAPI on a Tg(ubi:Loxp-stop-Loxp-hmga1a-eGFP, myl7:CreERT2) heart at 14 dpT. n = 6 hearts were analyzed. CM-specific nuclear Hmga1a–eGFP can be observed in most CMs. Scale bars, 20 μm. c, Representative images of immunofluorescent staining against Mef2, PCNA and Hmga1a–eGFP on 14-dpT control and hmga1a OE hearts. Arrowheads indicate proliferating CMs. Overview scale bars, 100 μm; zoom-in scale bars, 20 μm. d, Quantification of proliferating CMs in control (n = 5) and hmga1a OE (n = 6) hearts at 14 dpT. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test and show a significant difference between control and hmga1a OE (P = 0.0009). e, Workflow of tamoxifen treatment for long-term hmga1a OE used in f–k. dpf, days post fertilization; 4OH, 4-hydroxytamoxifen. f–j, Quantification of differences between control (n = 8) and hmga1a OE (n = 9) hearts, including myocardium-covered surface area (P = 0.0288) (f), total heart surface (myocardium + lumen) (P = 0.1721) (g), the percentage of total heart surface covered with myocardium (P < 0.001) (h), the percentage of proliferating CMs (P = 0.024) (i) and the density of cardiomyocyte nuclei (P = 0.2175) (j). Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed using two-tailed unpaired t-tests. k,l, Representative images of AFOG staining on a 1-year control and hmga1a OE zebrafish heart (k) and a 5-month nrg1 OE Tg(β-actin2:loxPmTagBFP-STOP-loxP-Nrg1) heart (l) showing muscle in orange, fibrin in red and collagen in blue. n = 8 control, n = 9 hmga1a OE and n = 1 nrg1 OE hearts were analyzed. Scale bars, 100 μm in the overviews and 50 μm in the zoom-ins. |
|
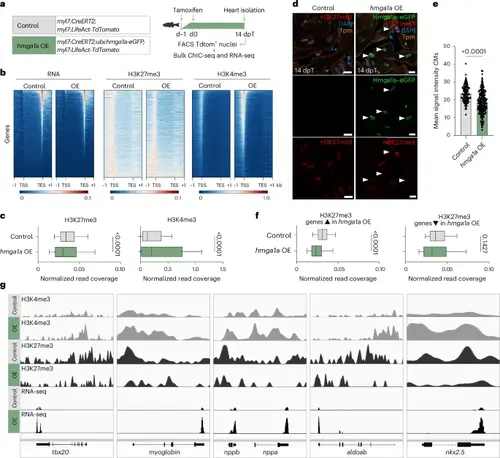
Reduction of repressive H3K27me3 marks by hmga1a OE in zebrafish CMs. a, Schematic overview for bulk sortChIC (control n = 10, hmga1a OE n = 10) and RNA-seq (control n = 14, hmga1a OE n = 14). b, Heatmap showing read distribution of RNA expression and levels of H3K27me3 and H3K4me3 in control and hmga1a OE CMs. TSS, transcription start site; TES, transcription end site; −1 and +1 indicate the kilobase distance from TSS or TES. c, Quantification of histone mark levels on all genes, comparing normalized read coverage in control versus hmga1a OE CMs. H3K27me3 levels (peak data on n = 21,440 genes) are significantly reduced on gene bodies (P < 0.001), and H3K4me3 levels (peak data on n = 21,843 genes) are significantly increased on promoter regions in hmga1a OE CMs (P < 0.001). Center line indicates median; whiskers indicate 10th/90th percentiles. Statistics were performed using two-tailed unpaired t-tests. d, Representative images of immunofluorescent staining for H3K27me3, tropomyosin (Tpm), Hmga1a–eGFP and DAPI on 14-dpT control and hmga1a OE hearts. Arrowheads indicate Hmga1a–eGFP+ CMs with low H3K27me3. Scale bars, 5 μm. e, Quantification of H3K27me3 signal intensity in single CMs of 14-dpT control (n = 8) versus hmga1a OE (n = 8) hearts. Datapoints represent single CM nuclei measured. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show a significant difference (P < 0.0001). f, Quantification of H3K27me3 levels on genes upregulated in hmga1a OE CMs (peak data on n = 1,141 genes) and genes downregulated in hmga1a OE CMs (peak data on n = 1,001 genes), comparing normalized read coverage in control versus hmga1a OE CMs. H3K27me3 levels were significantly reduced on gene bodies (P < 0.001) of genes upregulated in hmga1a OE CMs but not significantly different between control and hmga1a OE CMs on gene bodies of genes downregulated upon hmga1a OE. Center line indicates median; whiskers indicate 10th/90th percentiles. Statistics were performed using two-tailed unpaired t-tests. g, Genome tracks of example genes that were significantly upregulated in 14-dpT hmga1a OE CMs and were found downstream of Hmga1a (modules 5–8 in the scRNA-seq). |
|
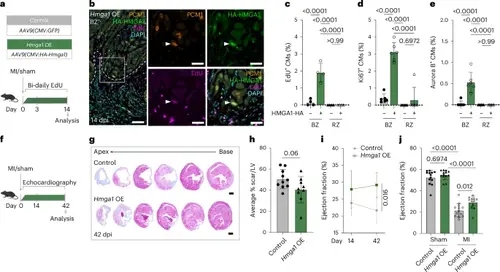
HMGA1 promotes CM proliferation and cardiac regeneration in injured adult mice. a, Schematic overview for experiments in b–e. b, Representative image of immunofluorescent staining against PCM-1, HA and EdU. Dashed line indicates the injury border. Arrowheads indicate HMGA1-HA+EdU+ CMs. Scale bars, 100 μm in overview and 20 μm in zoom-in. c–e, Quantification of EdU+ (c), Ki67+ (d) and Aurora B+ (e) CMs within the BZ and RZ of hearts transduced with HA-HMGA1. n = 4 hearts were analyzed for EdU and n = 6 for Ki67/Aurora B quantification. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show significant differences for % EdU+/Ki67+/AuroraB+ CMs in HMGA1-HA+ BZ CMs compared to HA− BZ CMs and RZ HA+/− CMs (P < 0.0001 for all). No significant difference was found between RZ HA− and HA+ cells for % EdU+ CMs (P > 0.99), % Ki67+ CMs (P = 0.6972) and % AuroraB+ CMs (P > 0.99). f, Workflow for mouse experiments in g–j. g, Representative images of control and Hmga1 OE hearts at 42 dpi stained with Masson’s trichrome. Distance between sections is 400 μm. Scale bars, 1 mm. h, Quantification of scar size in control (n = 10) and Hmga1 OE (n = 9) hearts at 42 dpi showing average % MI length/midline LV length. Error bars indicate mean ± s.d. Statistics were performed by two-tailed unpaired t-test and show no significant difference (P = 0.06). i, Quantification of EF at 14 dpi and 42 dpi of control (n = 13) and Hmga1 OE (n = 13) hearts. Error bars indicate mean ± s.d. Statistics were performed using a two-way ANOVA followed by Sidak’s multiple comparisons test and show a significant difference (P = 0.016). j, Quantification of EF at 42 dpi of sham (n = 13 control, n = 13 Hmga1 OE) and MI (n = 13 control, n = 12 Hmga1 OE) hearts. Datapoints represent individual hearts. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show a significant difference between control sham/MI hearts (P < 0.0001), between Hmga1 OE sham/MI hearts (P < 0.0001) and between control and Hmga1 OE MI hearts (P = 0.01) but not between control and Hmga1 OE sham hearts (P = 0.6974). |
|
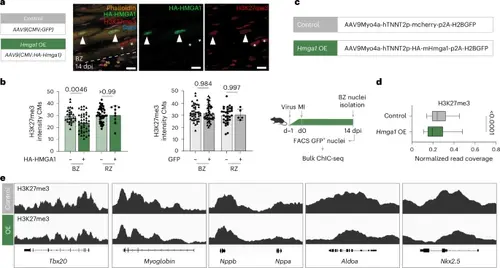
Reduction of repressive H3K27me3 marks by HMGA1 in mouse BZ CMs. a, Schematic overview of experiments (a,b) and representative image of immunofluorescent staining against H3K27me3, HMGA1-HA, phalloidin and DAPI on 14-dpi mouse hearts transduced with AAV9(CMV:HA-Hmga1). Dashed line indicates the injury border. Arrowheads indicate HMGA1-HA transduced CMs; asterisks indicate non-transduced CMs. Scale bar, 5 μm. b, Quantification of H3K27me3 signal intensity in single CMs in the BZ and RZ of 14-dpi Hmga1 (n = 4) and GFP (n = 4) virus transduced hearts. Datapoints represent single CM nuclei measured. Error bars indicate mean ± s.d. Statistics were performed using a one-way ANOVA followed by Tukey’s multiple comparisons test and show a significant difference between HA+ BZ CMs and HA− BZ CMs (P = 0.0046). c, Schematic overview for bulk sortChIC on Hmga1 OE CMs (c–e). d, Quantification of H3K27me3 levels on all genes, comparing normalized read coverage in control versus Hmga1 OE BZ CMs. H3K27me3 levels (peak data on n = 21,937 genes) are significantly reduced on gene bodies (P < 0.0001) in Hmga1 OE BZ CMs. Center line indicates median; whiskers indicate 10th/90th percentiles. Statistics were performed using a two-tailed unpaired t-test. e, Genome tracks of orthologs to example genes in Fig. 6g that are significantly upregulated in 14-dpT hmga1a OE CMs and were found downstream of Hmga1a (modules 5–8 in the scRNA-seq). Tracks show reduced H3K27me3 levels on genes in Hmga1 OE BZ CMs. |