- Title
-
Embryo-scale reverse genetics at single-cell resolution
- Authors
- Saunders, L.M., Srivatsan, S.R., Duran, M., Dorrity, M.W., Ewing, B., Linbo, T.H., Shendure, J., Raible, D.W., Moens, C.B., Kimelman, D., Trapnell, C.
- Source
- Full text @ Nature
|
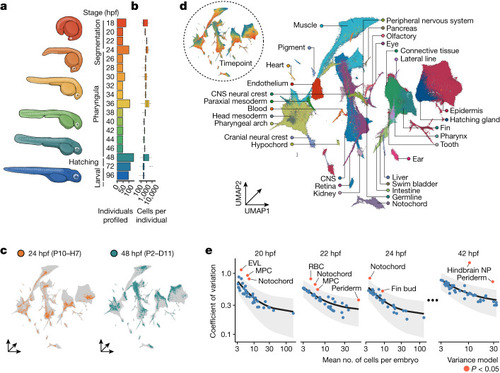
Collection of an individual-resolved single-cell zebrafish atlas using oligonucleotide hashing. |
|
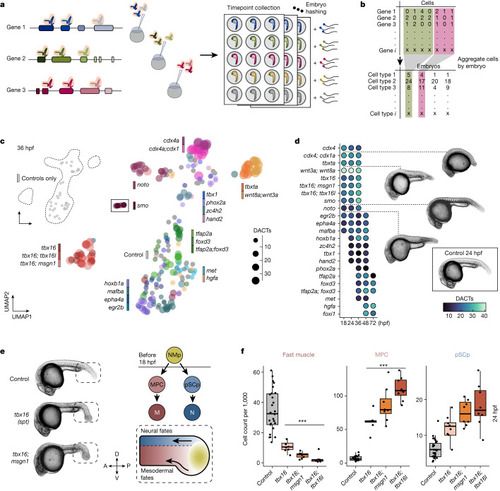
High-resolution phenotyping of crispant zebrafish embryos. |
|
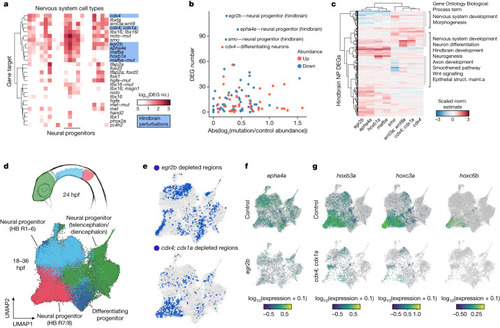
Systematic detection of DEGs and cell state changes across perturbations. |
|
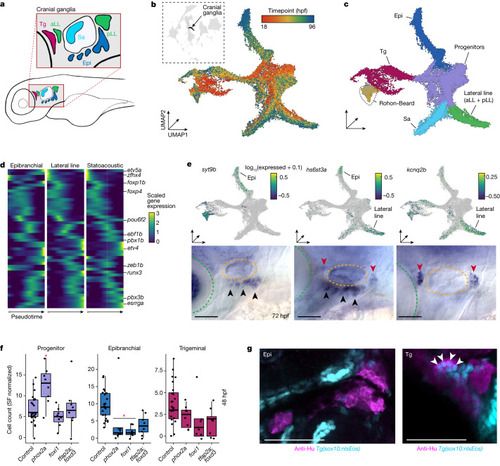
Whole-embryo phenotyping robustly captures effects in cranial sensory neurons. |
|
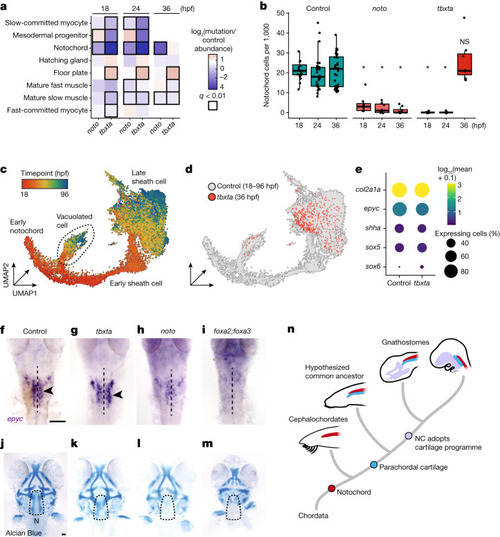
Tbxta and Noto perturbations uncover the genetic requirements of cranial cartilage development. |