- Title
-
Loss of Sec-1 Family Domain-Containing 1 (scfd1) Causes Severe Cardiac Defects and Endoplasmic Reticulum Stress in Zebrafish
- Authors
- Huttner, I.G., Santiago, C.F., Jacoby, A., Cheng, D., Trivedi, G., Cull, S., Cvetkovska, J., Chand, R., Berger, J., Currie, P.D., Smith, K.A., Fatkin, D.
- Source
- Full text @ J Cardiovasc Dev Dis
|
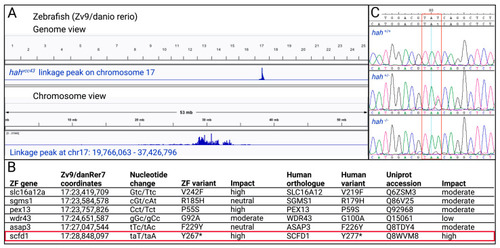
Mapping of zebrafish (ZF) |
|
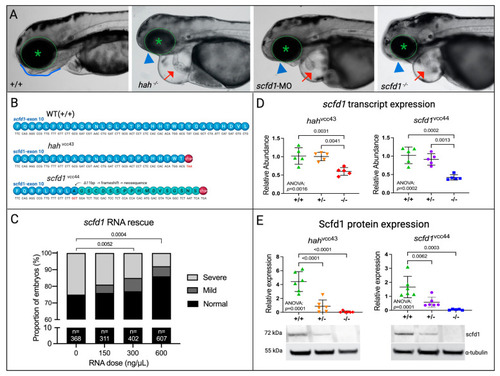
EXPRESSION / LABELING:
PHENOTYPE:
|
|
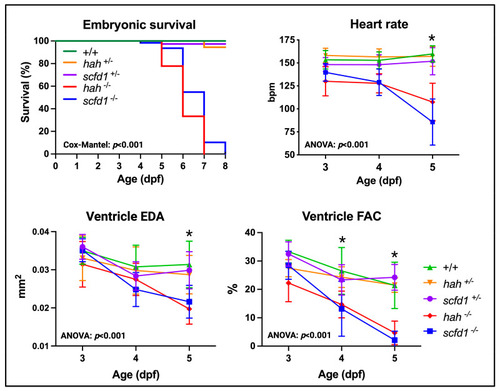
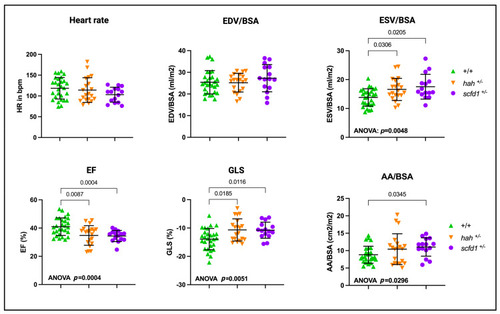
Scfd1 deficiency leads to cardiac dysfunction in zebrafish embryos. Survival, heart rate, ventricular end-diastolic area (EDA) and fractional area change (FAC) are reduced in embryonic |
|
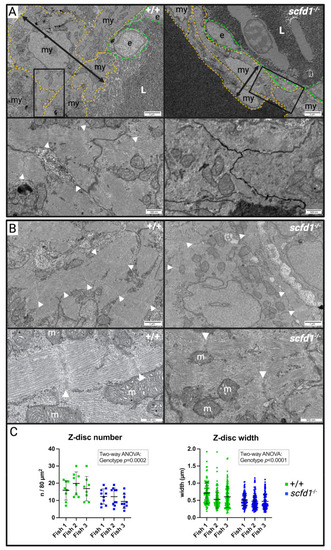
Homozygous PHENOTYPE:
|
|
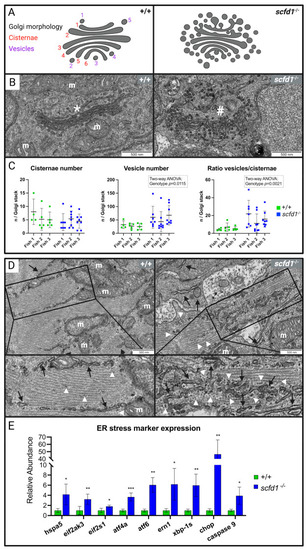
Altered cardiomyocyte Golgi apparatus and reticular network morphology, and upregulation of endoplasmic reticulum (ER) stress markers in EXPRESSION / LABELING:
PHENOTYPE:
|
|
PHENOTYPE:
|