- Title
-
Ligament injury in adult zebrafish triggers ECM remodeling and cell dedifferentiation for scar-free regeneration
- Authors
- Anderson, T., Mo, J., Gagarin, E., Sherwood, D., Blumenkrantz, M., Mao, E., Leon, G., Levitz, H., Chen, H.J., Tseng, K.C., Fabian, P., Crump, J.G., Smeeton, J.
- Source
- Full text @ NPJ Regen Med
|
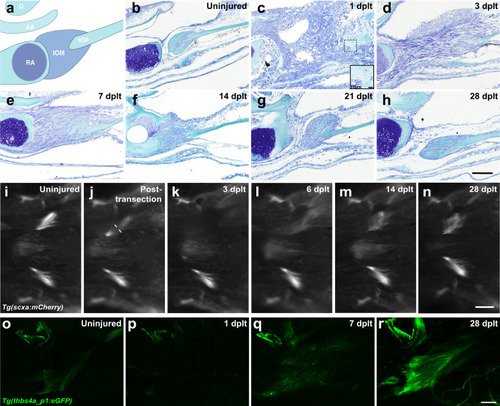
Adult zebrafish ligament transection injury and regeneration. |
|
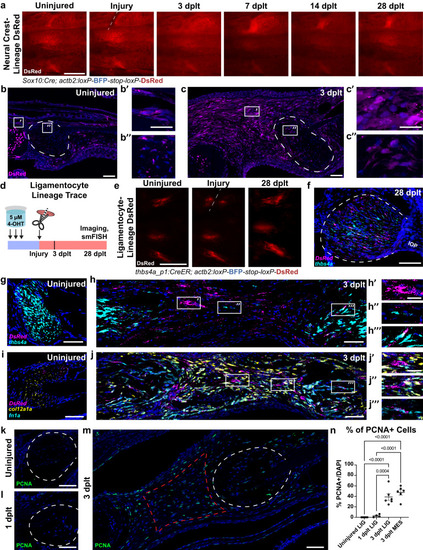
Lineages contributing to ligament regeneration following transection injury. |
|
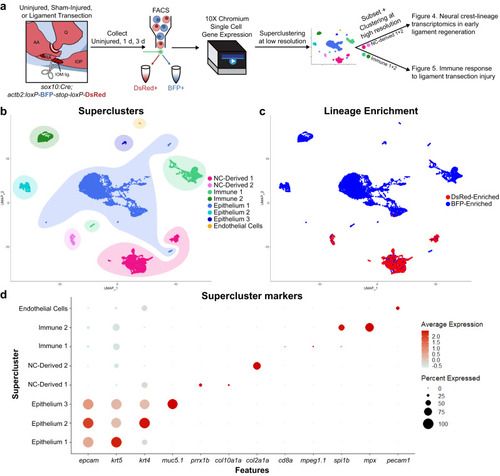
Single-cell transcriptomics of joint tissue through early ligament regeneration. |
|
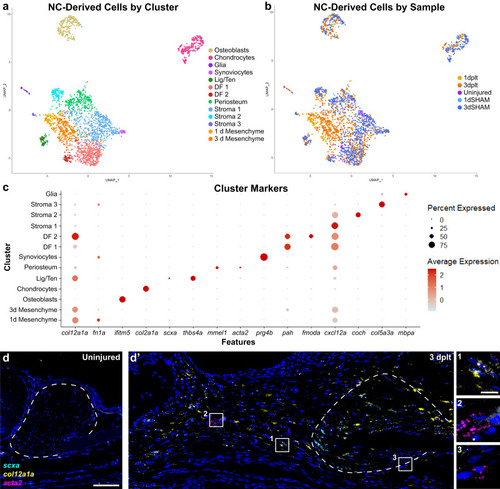
Neural crest-lineage single-cell transcriptomics in early ligament regeneration. |
|
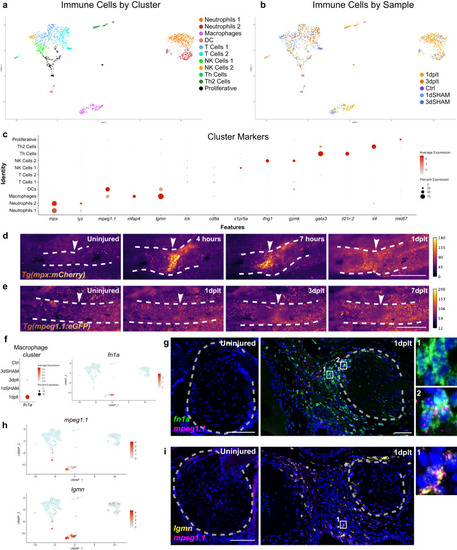
Immune response to ligament transection injury. UMAPs of single-cell RNA sequencing datasets colored by cluster ( |
|
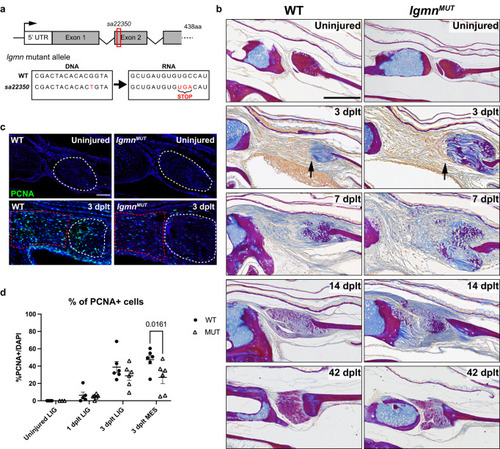
Dysregulated ECM remodeling and decreased proliferation in |
|
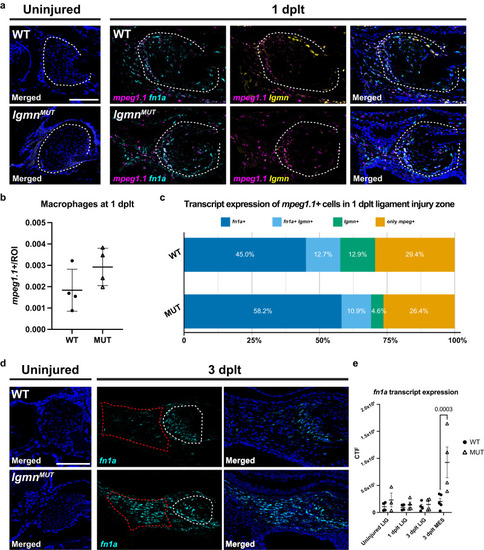
Abnormal expression of ECM factors in |
|
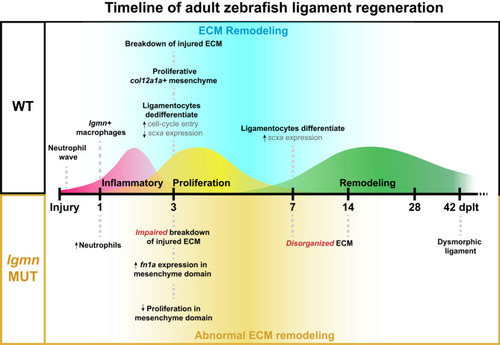
Adult zebrafish ligament regeneration timeline and major milestones. Schematic timeline of ligament regeneration in wild type and |