- Title
-
Generation and Characterization of a Zebrafish Model for ADGRV1-Associated Retinal Dysfunction Using CRISPR/Cas9 Genome Editing Technology
- Authors
- Stemerdink, M., Broekman, S., Peters, T., Kremer, H., de Vrieze, E., van Wijk, E.
- Source
- Full text @ Cells
|
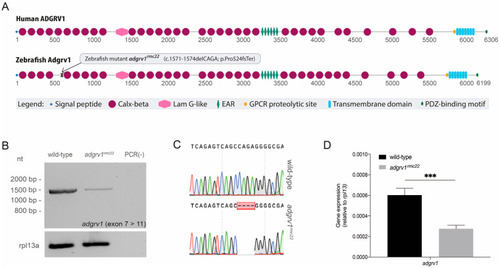
Domain architecture of human and zebrafish ADGRV1 and transcript analysis in homozygous |
|
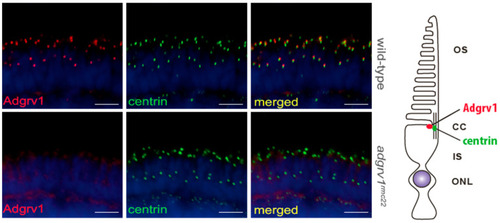
Localization of Adgrv1 in retinal cryosections of wild-type and |
|
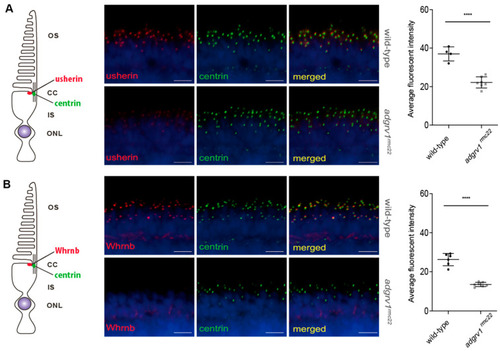
Reduced expression of usherin and Whrnb at the photoreceptor periciliary region of |
|
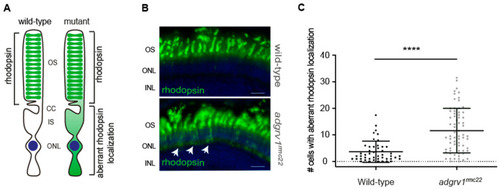
Aberrant localization of rhodopsin in photoreceptor cell bodies in the |
|
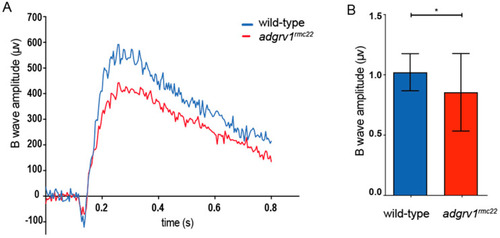
Electroretinogram recordings reveal impaired retinal function in PHENOTYPE:
|