- Title
-
The Zinc Transporter SLC39A10 Plays an Essential Role in Embryonic Hematopoiesis
- Authors
- He, X., Ge, C., Xia, J., Xia, Z., Zhao, L., Huang, S., Wang, R., Pan, J., Cheng, T., Xu, P.F., Wang, F., Min, J.
- Source
- Full text @ Adv Sci (Weinh)
|
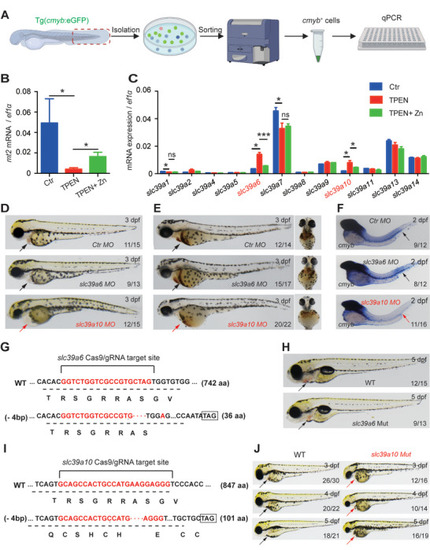
Functional screening in zebrafish reveals Sslc39a10 as a potentially important zinc transporter in early hematopoiesis. A) Schematic diagram depicting the strategy for performing functional screening in zebrafish. B,C) Relative levels of the indicated |
|
|
|
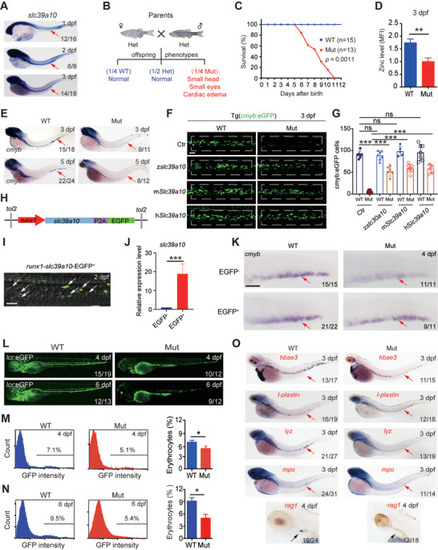
Loss of |
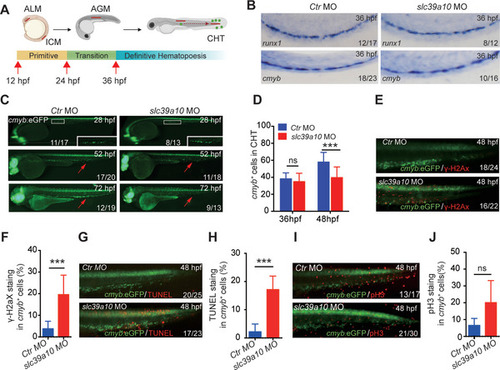
|
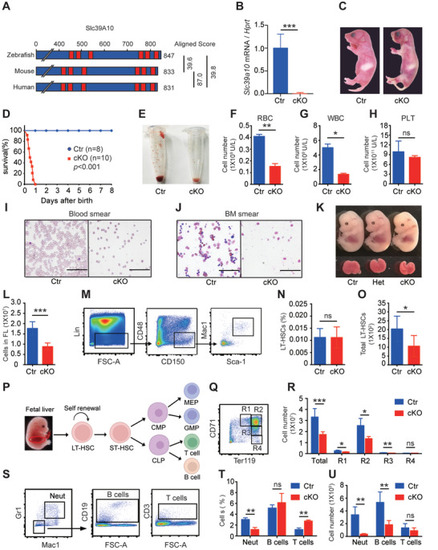
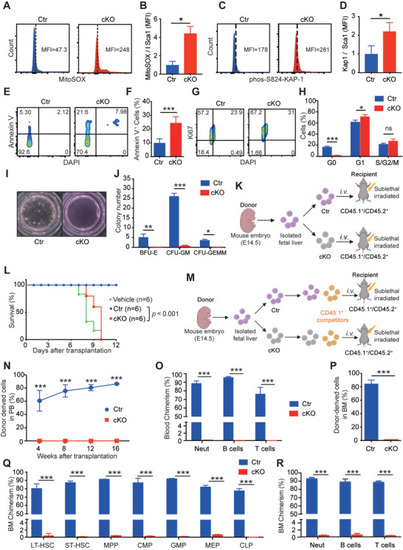
Mice lacking |
|
|
|
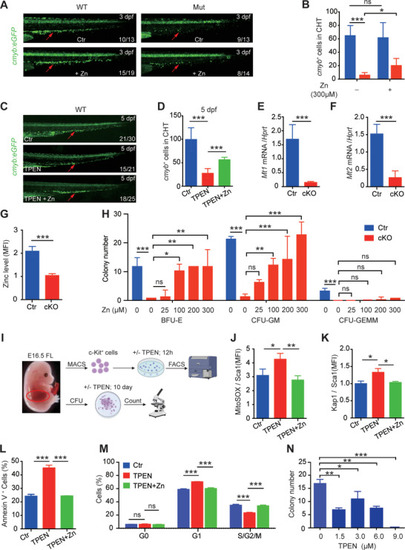
Zinc supplementation significantly reduces the impaired properties of HSPCs in |
|
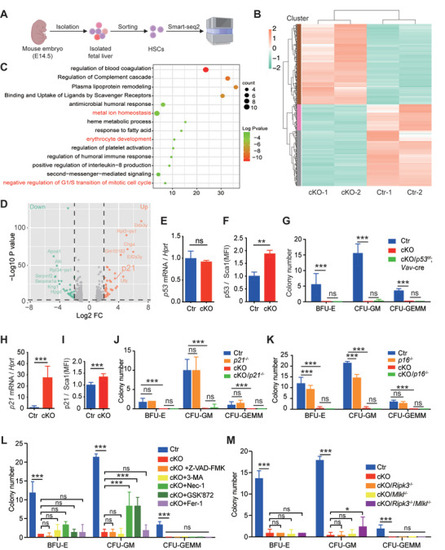
Inhibiting necroptosis partially restores the colony‐forming capacity of |