- Title
-
Probiotics Enhance Bone Growth and Rescue BMP Inhibition: New Transgenic Zebrafish Lines to Study Bone Health
- Authors
- Sojan, J.M., Raman, R., Muller, M., Carnevali, O., Renn, J.
- Source
- Full text @ Int. J. Mol. Sci.
|
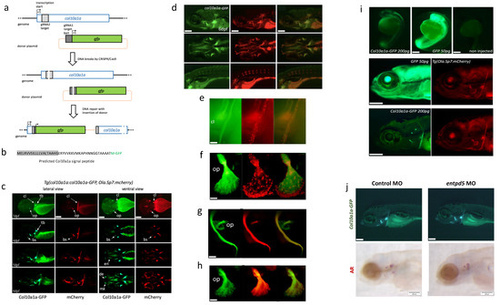
Figure 1. Characterisation of the Tg(col10a1a:col10a1a-GFP) transgenic line. (a) Schematic representation of the endogenous col10a1a gene (top line), the plasmids used for microinjection along with the two gRNAs (bait and col10a1a, respectively gRNA1 and gRNA2), the resulting cuts in the genomic DNA and plasmids, and the expected reporter construct in the transgenic genome. (b) N-terminal end of the fusion protein produced by the Tg(col10a1a:col10a1-GFP) transgenic line, with the predicted signal peptide in grey and the original GFP translational start site (M) in green. (c) Timeline of expression of GFP (green fluorescence) and mCherry (red fluorescence) in double-transgenic larvae Tg(col10a1a:col10a1a-GFP; Ola.Sp7:mCherry). Lateral and ventral views at different developmental stages as indicated, anterior to the left. White arrows point to specific elements: (bs) branchiostegal ray, (cl) cleithrum, (de) dentary, (en) entopterygoid, (mx) maxillary, (op) opercle and (tb) tooth bud. (d) Expression of Col10a1a-GFP protein (green) in 6 dpf Tg(col10a1a:col10a1a-GFP) larvae live-stained with AR to visualize mineralized bone. (e–h) Close inspection of Col10a1a-GFP localization (green) compared to mCherry expression by osteoblasts in (e) the cleithrum (cl) or the opercle (f) of 9 dpf Tg(col10a1a:col10a1a-GFP; Ola.Sp7:mCherry) zebrafish larvae. (g,h) Expression of GFP protein (green) in Tg(col10a1a:col10a1a-GFP) 6 dpf larvae live-stained with AR to visualize bone matrix. Close inspection of the cleithrum (g) and the opercle (h). (i) Zebrafish Tg(Ola.Sp7:mCherry) larvae after microinjection of mRNA coding for GFP or for the fusion protein Col10a1a-GFP. Top: embryos at 1 dpf, showing weak fluorescence of Col10a1a-GFP, extremely strong fluorescence of GFP, and no fluorescence in controls. Bottom: the same larvae at 5 dpf, still showing strong GFP expression in the entire body and weak, but specific fluorescence of Col10a1a-GFP located in bone elements (cleithrum and opercle) as confirmed by the red fluorescence of osteoblast-specific mCherry. (j) Morpholino injection into Tg(col10a1a:col10a1a-GFP) larvae. The Col10a1a-GFP protein labels cranial bone elements at 4 dpf in both control and entpd5 MO injected larvae (top), but AR staining is absent in entpd5 morphants (bottom). The scale bars, given in the left bottom corner of the images, represent 200 µm, except for (e–h), where they indicate 20 µm. EXPRESSION / LABELING:
|
|
Figure 2. GFP expression in the Tg(sp7:sp7-GFP) transgenic line. (a) N-terminal end of the fusion protein produced by the Tg(sp7:sp7-GFP) transgenic line, with the predicted signal peptide in grey and the original GFP translational start site (M) in green. (b) Transgene expression in the Tg(sp7:sp7-GFP) and the previously described Tg(Ola.Sp7:mCherry) lines tracked in the head region (top: lateral view and bottom: ventral view) from 3 dpf to 10 dpf. Earlier transgene expression occurs in the maxillary (white arrows) and pharyngeal tooth buds (white arrowheads) at 3 dpf and in the entopterygoid (yellow arrows) at 6 dpf of the Tg(sp7:sp7-GFP) line. The scale bars, given in the left bottom corner of the images in the top row, correspond to 100 µm.
EXPRESSION / LABELING:
|
|
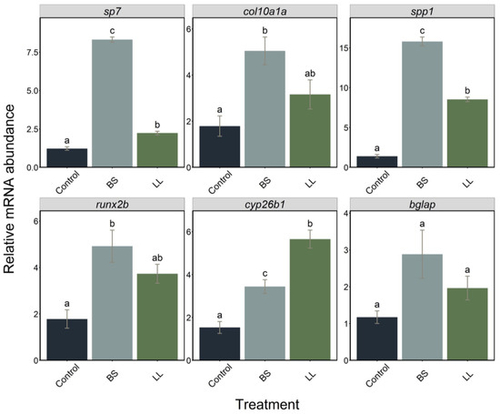
Figure 3. Relative expression levels of sp7, col10a1a, spp1, runx2b, cyp26b1 and bglap genes in larvae (n = 7) treated with two probiotics, BS and LL, sampled at 7 dpf. Data are presented as mean ± S.D. One-way ANOVA and Tukey’s multiple comparison tests are used. Different letters denote statistically significant differences (p < 0.05) between the experimental groups.
|
|
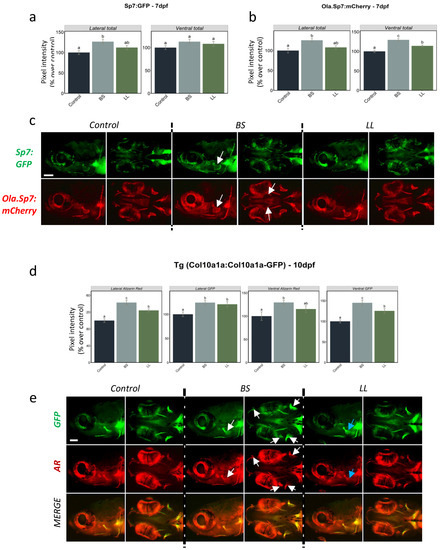
Figure 4. Integrated pixel intensity values for various areas measured in 7 dpf zebrafish from (a) Tg(sp7:sp7-GFP) and (b) Tg(Ola.Sp7:mCherry) larvae in controls and upon two different probiotic treatments. (c) Signal expression images (lateral and ventral views) of the head area of Tg(sp7:sp7-GFP) and Tg(Ola.Sp7:mCherry) larvae under the different conditions. (d) Integrated pixel intensity values for various areas measured in 10 dpf Tg(col10a1a:col10a1a-GFP) zebrafish larvae from the different treatment groups and stained with AR. (e) GFP, AR fluorescence and merged images of the head area (lateral and ventral views) of Tg(col10a1a:col10a1a-GFP) larvae from the different treatment groups. Increased GFP and AR fluorescence in various bony structures are denoted by white and blue arrows, respectively, in BS- and LL-treated fish. One-way ANOVA and Tukey’s multiple comparison tests are used, and statistical significance was set at p < 0.05. Different letters denote statistically significant differences between experimental groups. The scale bars, given in the left bottom corner of the first image in the top row, correspond to 100 µm.
|
|
Figure 5. Effects of LDN212854 on sp7 expression. Integrated pixel intensity values (mCherry red fluorescence) for 5 dpf Tg(Ola.Sp7:mCherry) larvae measured in ventral view treated with 10 µM and 20 µM LDN212854 from 2 dpf to 4 dpf. Different letters denote statistically significant differences between experimental groups (one-way ANOVA, p < 0.05, followed by Tukey’s post hoc test).
|
|
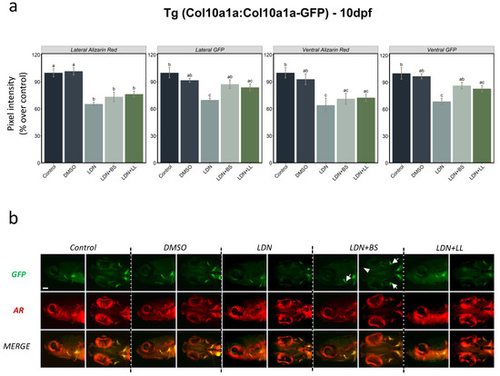
Figure 6. BMP inhibitor exposure followed by two probiotics treatments in Tg(col10a1a:col10a1a-GFP) larvae. (a) Integrated pixel intensity values for various areas measured in 10 dpf zebrafish Tg(col10a1a:col10a1a-GFP) larvae divided into various groups—control, DMSO, LDN, LDN+BS and LDN+LL—and stained with AR. DMSO was used as additional control since it was employed as the solvent for LDN. One-way ANOVA and Tukey’s multiple comparison tests were used, and statistical significance was set at p < 0.05. Different letters denote statistically significant differences among the experimental groups. (b) GFP, AR staining and merged images of the head area (lateral and ventral views) of Tg(col10a1a:col10a1a-GFP) larvae of the different treated groups. White arrows denote GFP in various bony structures and the white arrowhead indicates the presence of a signal in additional structures (that was absent in other groups) in BS-treated fish after LDN exposure (LDN + BS).
|