- Title
-
Starvation causes changes in the intestinal transcriptome and microbiome that are reversed upon refeeding
- Authors
- Jawahar, J., McCumber, A.W., Lickwar, C.R., Amoroso, C.R., de la Torre Canny, S.G., Wong, S., Morash, M., Thierer, J.H., Farber, S.A., Bohannan, B.J.M., Guillemin, K., Rawls, J.F.
- Source
- Full text @ BMC Genomics
|
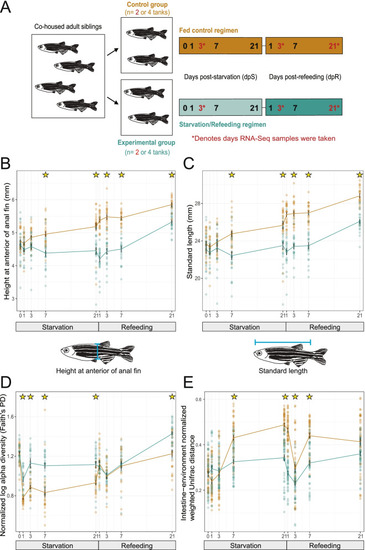
Starvation and refeeding affect zebrafish somatic growth as well as intestinal and environmental microbiome diversity. |
|
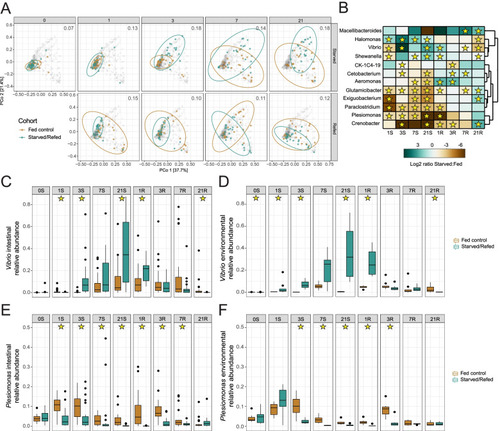
Starvation and refeeding dynamically alters composition of the adult zebrafish intestinal microbiome. |
|
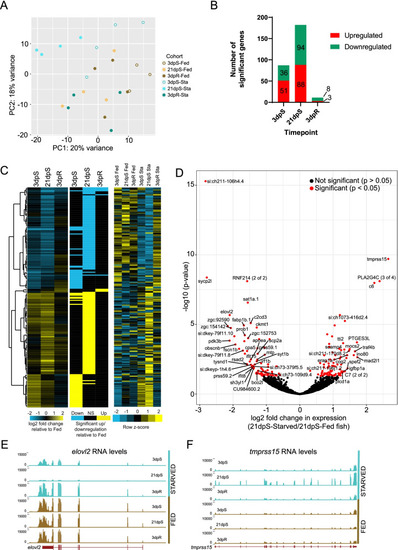
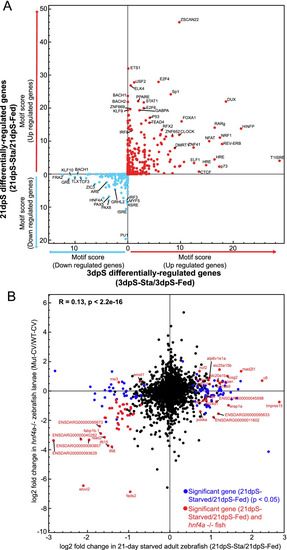
Starved zebrafish differentially regulate intestinal gene expression when compared to fed zebrafish. |
|
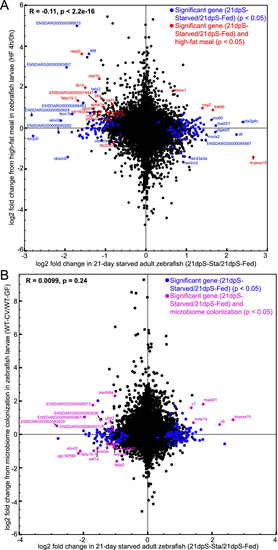
Some genes responsive to starvation in the intestine are also responsive to high fat feeding and microbial colonization. |
|
The transcription factor |