- Title
-
The Physical Role of Mesenchymal Cells Driven by the Actin Cytoskeleton Is Essential for the Orientation of Collagen Fibrils in Zebrafish Fins
- Authors
- Kuroda, J., Itabashi, T., Iwane, A.H., Aramaki, T., Kondo, S.
- Source
- Full text @ Front Cell Dev Biol
|
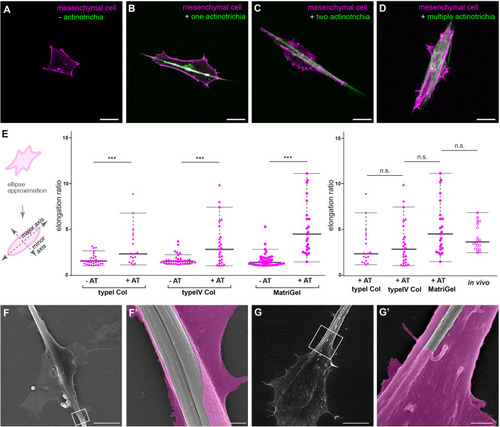
Distribution pattern of the mesenchymal cells in larval fins. |
|
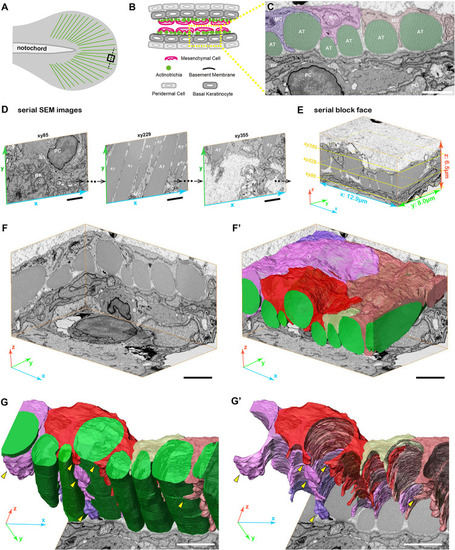
3D reconstruction of fin mesenchymal cells and actinotrichia by FIB-SEM analysis. |
|
Interaction between mesenchymal cells and actinotrichia |
|
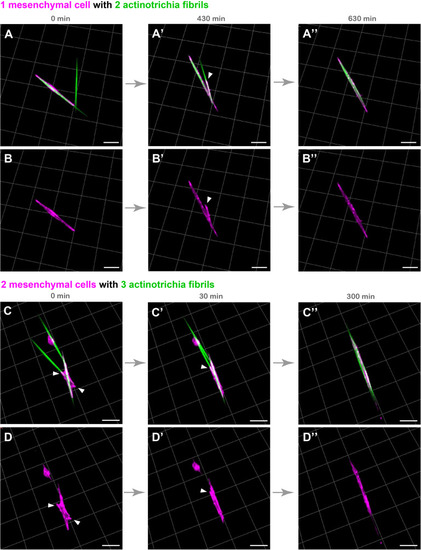
Live cell imaging of cultured mesenchymal cells holding the actinotrichia fibrils. Captured time-lapse images for the interaction between mesenchymal cells and actinotrichia fibrils. Mesenchymal cells and actinotrichia were isolated from the fins of F1 TG larvae [TG; 5x |
|
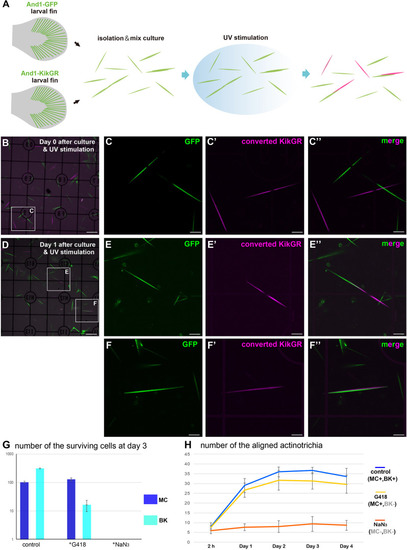
Mesenchymal cells are essential for the |
|
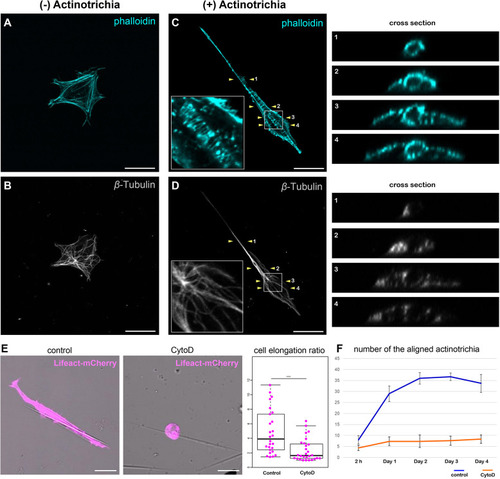
Inhibition of actin polymerization suppressed the orientation formation of actinotrichia. |
|
Suppression of actin polymerization in mesenchymal cells induced the collapse of the actinotrichia distribution |