- Title
-
Targeted RNA Knockdown by a Type III CRISPR-Cas Complex in Zebrafish
- Authors
- Fricke, T., Smalakyte, D., Lapinski, M., Pateria, A., Weige, C., Pastor, M., Kolano, A., Winata, C., Siksnys, V., Tamulaitis, G., Bochtler, M.
- Source
- Full text @ CRISPR J
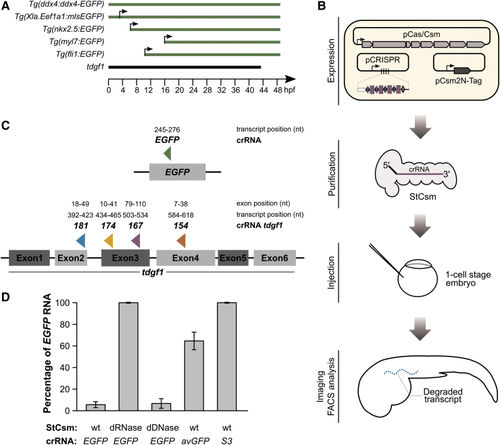
|
Experimental design and |
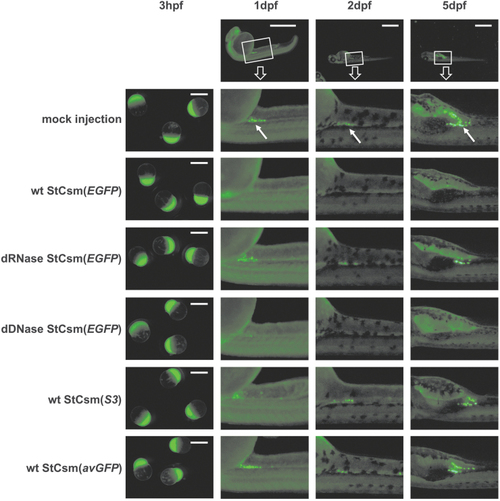
|
Microscopy of StCsm mediated |
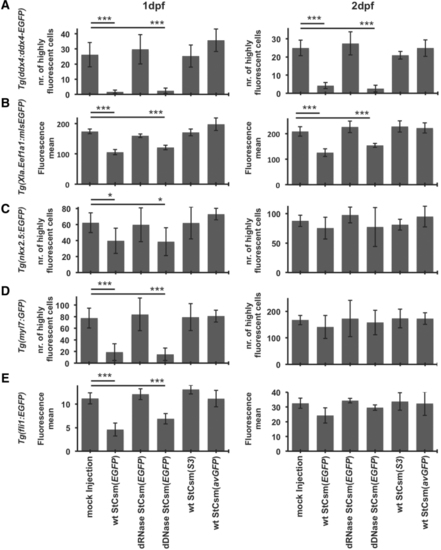
|
Quantification of |
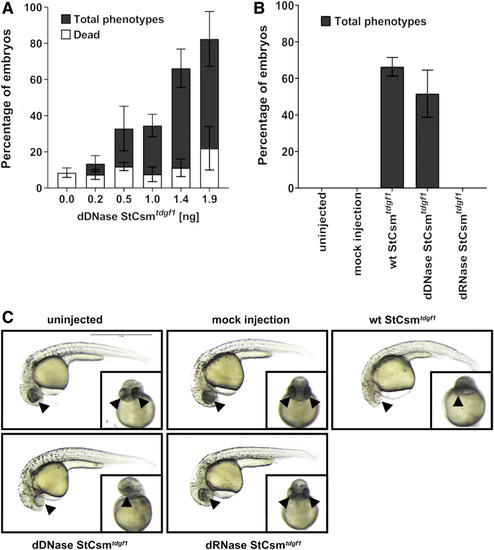
|
Knockdown of endogenous |
|
Analysis of on-target effects of |
|
Analysis of off-target effects of |