- Title
-
Loss of Caveolin-1 and caveolae leads to increased cardiac cell stiffness and functional decline of the adult zebrafish heart
- Authors
- Grivas, D., González-Rajal, Á., Guerrero Rodríguez, C., Garcia, R., de la Pompa, J.L.
- Source
- Full text @ Sci. Rep.
|
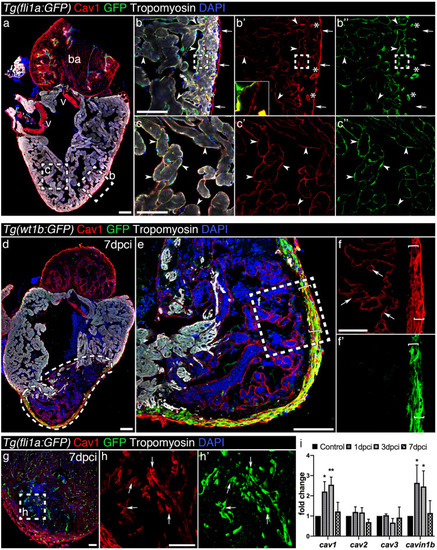
Caveolin-1 is expressed in the endothelium, endocardium and epicardium of the intact and injured adult zebrafish heart. ( |
|
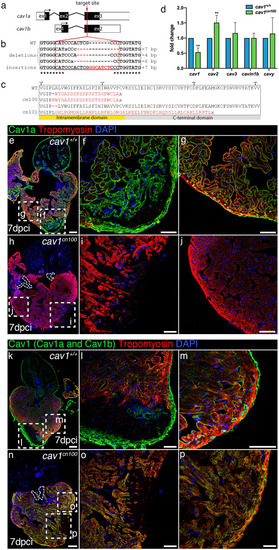
Generation of |
|
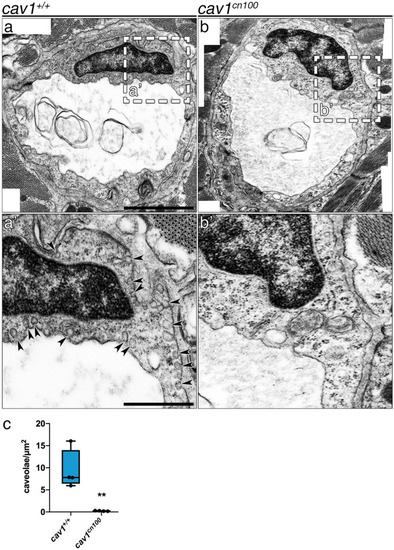
Loss of caveolae in PHENOTYPE:
|
|
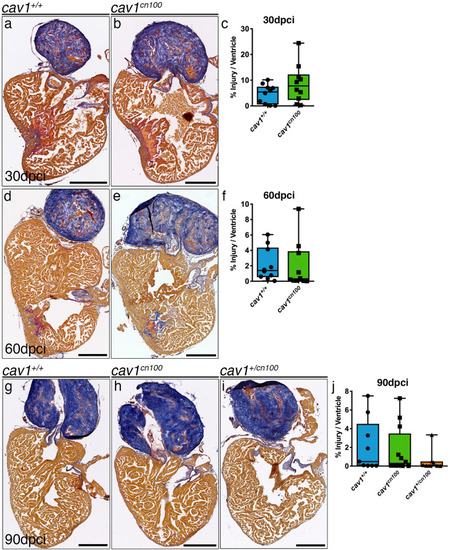
Heart regeneration is unaffected in PHENOTYPE:
|
|
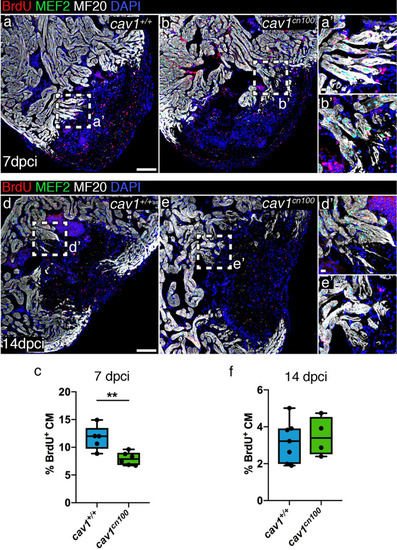
Cardiomyocyte proliferation is transiently reduced upon cryoinjury in |
|
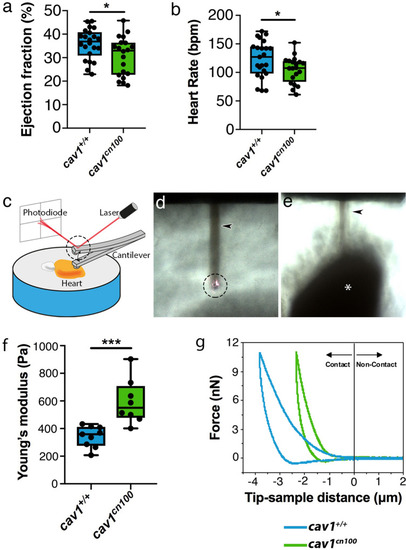
Impaired cardiac function and stiffer heart tissue in caveolae-deprived PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|