- Title
-
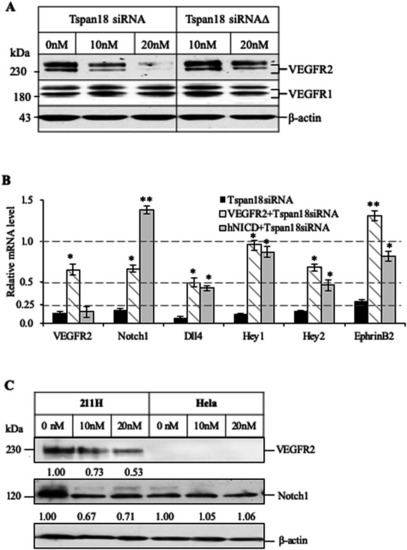
Tetraspanin18 regulates angiogenesis through VEGFR2 and Notch pathways
- Authors
- Li, G.X., Zhang, S., Liu, R., Singh, B., Singh, S., Quinn, D.I., Crump, G., Gill, P.S.
- Source
- Full text @ Biol. Open
|
|
|
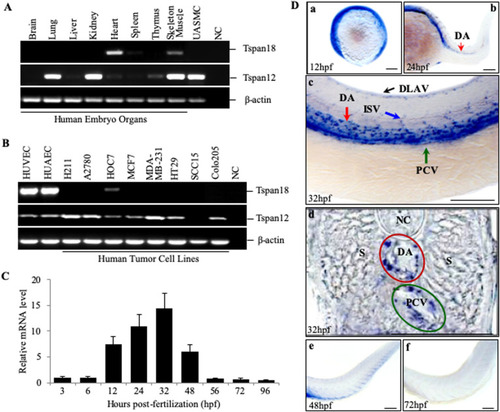
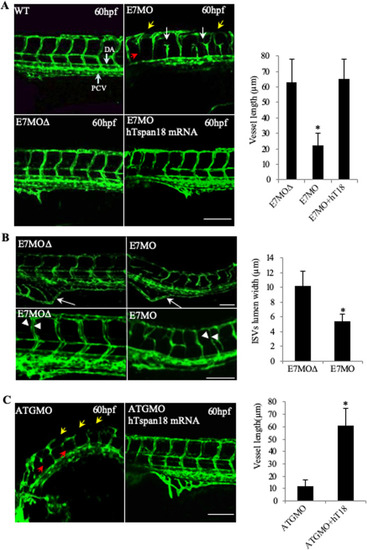
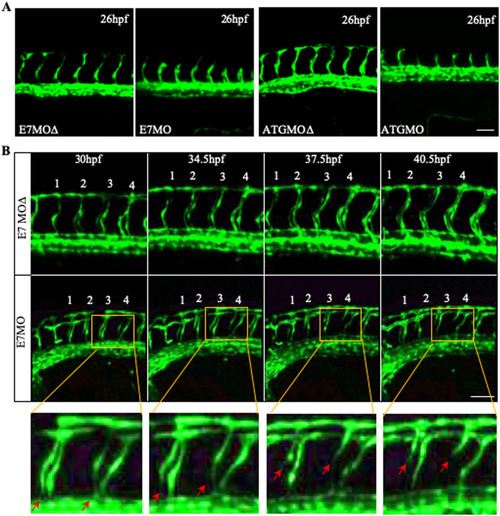
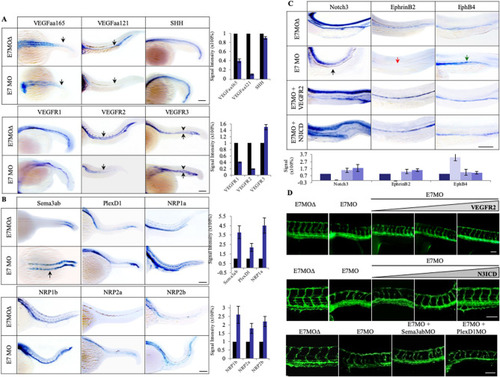
Vascular defects in Tspan18-zebrafish morphants. (A) Morphological and vascular defects were examined in E7MO-injected zebrafish at 60 hpf. E7MO represents the morpholino targeting Tspan18 Exon 7 splicing site. E7MOΔ is a control morpholino with five-base mismatch. WT represents fish injected with morpholino diluent only. Arrows in the picture of Tspan18-deficient fish (E7MO) point to missing ISV (red), ectopic ISV branching (white) or discontinuous DLAV (yellow). Quantification of ISV length of E7MO and E7MOΔ injected fish (n=80) are shown on the bottom left. Co-injection of a human sense capped Tspan18 mRNA rescued the defects caused by E7MO. (B) E7MOΔ injected embryos had reduced subintestinal vessels (upper panel, arrows) and narrow ISV lumen (lower panel, arrowheads) at 72 hpf. Quantification of ISV lumen width from each group is shown on the right (n=75). (C) Morphological and vascular defects were examined in ATGMO injected zebrafish at 60 hpf. ATGMO represents the morpholino targeting Tspan18 ATG start codon containing Exon 3 splicing site. Arrows are designated as in A. Co-injection of a human sense capped Tspan18 mRNA rescued the defects caused by ATGMO. Error bars represent s.d. P value was calculated using two-tailed Student's t-test. *P<0.02. All larvae lateral view. Anterior is to the left. Scale bars: 50 μm. All experiments (A–C) were repeated at least three times and similar results were obtained. PHENOTYPE:
|
|
|
|
|
|
|
|
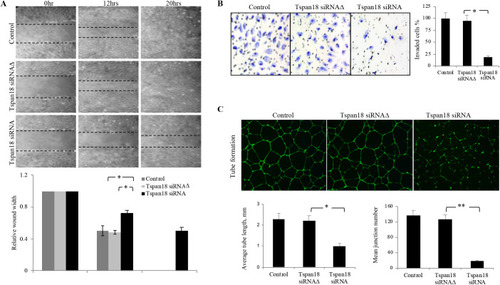
Tspan18 knockdown inhibits angiogenesis in vitro. (A) Tspan18 knockdown inhibited migration of HUAEC in an in vitro wound-healing assay. Transfection reagents served as a negative control. The margin of the wound is demarcated transwelled lines. Three independent experiments were performed and quantified wound width is shown (normalized to 0 h width). (B) Tspan18 knockdown inhibited invasion of HUAEC in transwell invasion assay. Quantification of invaded cells is shown on the right. (C) Tspan18 knockdown inhibited HUAEC tube formation on Matrigel. Quantification of tube length and number of junctions is shown on the bottom. The experiments in A–C were repeated three times with similar results. Error bars: s.d., *P<0.01, **P<0.002. P values were calculated using a two-tailed Student's t-test. |
|
|