- Title
-
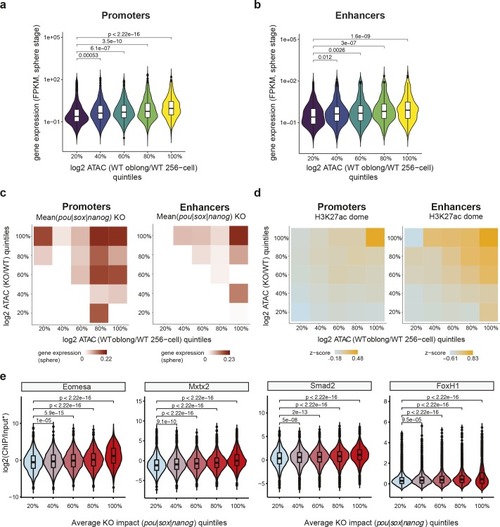
Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation
- Authors
- Pálfy, M., Schulze, G., Valen, E., Vastenhouw, N.L.
- Source
- Full text @ PLoS Genet.
|
|
|
|
|
|
|
PHENOTYPE:
|
|
|