- Title
-
Loss of UGP2 in brain leads to a severe epileptic encephalopathy, emphasizing that bi-allelic isoform-specific start-loss mutations of essential genes can cause genetic diseases
- Authors
- Perenthaler, E., Nikoncuk, A., Yousefi, S., Berdowski, W.M., Alsagob, M., Capo, I., van der Linde, H.C., van den Berg, P., Jacobs, E.H., Putar, D., Ghazvini, M., Aronica, E., van IJcken, W.F.J., de Valk, W.G., Medici-van den Herik, E., van Slegtenhorst, M., Brick, L., Kozenko, M., Kohler, J.N., Bernstein, J.A., Monaghan, K.G., Begtrup, A., Torene, R., Al Futaisi, A., Al Murshedi, F., Mani, R., Al Azri, F., Kamsteeg, E.J., Mojarrad, M., Eslahi, A., Khazaei, Z., Darmiyan, F.M., Doosti, M., Karimiani, E.G., Vandrovcova, J., Zafar, F., Rana, N., Kandaswamy, K.K., Hertecant, J., Bauer, P., AlMuhaizea, M.A., Salih, M.A., Aldosary, M., Almass, R., Al-Quait, L., Qubbaj, W., Coskun, S., Alahmadi, K.O., Hamad, M.H.A., Alwadaee, S., Awartani, K., Dababo, A.M., Almohanna, F., Colak, D., Dehghani, M., Mehrjardi, M.Y.V., Gunel, M., Ercan-Sencicek, A.G., Passi, G.R., Cheema, H.A., Efthymiou, S., Houlden, H., Bertoli-Avella, A.M., Brooks, A.S., Retterer, K., Maroofian, R., Kaya, N., van Ham, T.J., Barakat, T.S.
- Source
- Full text @ Acta Neuropathol.
|
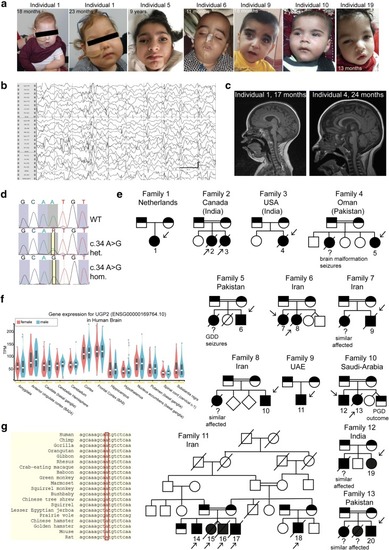
UGP2 homozygous variants in 20 individuals with severe epileptic encephalopathy. |
|
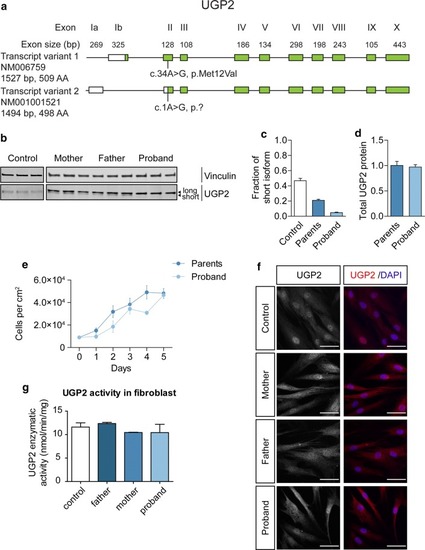
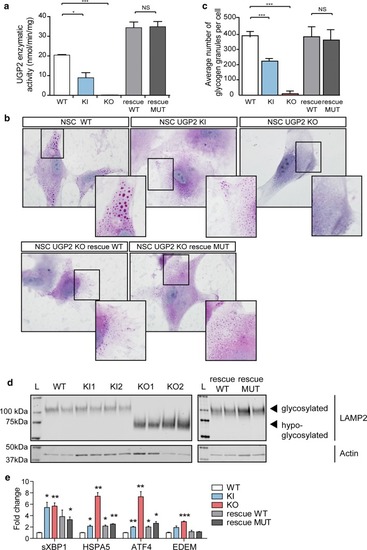
UGP2 homozygous variant leads to a loss of the shorter protein isoform in patient fibroblasts. |
|
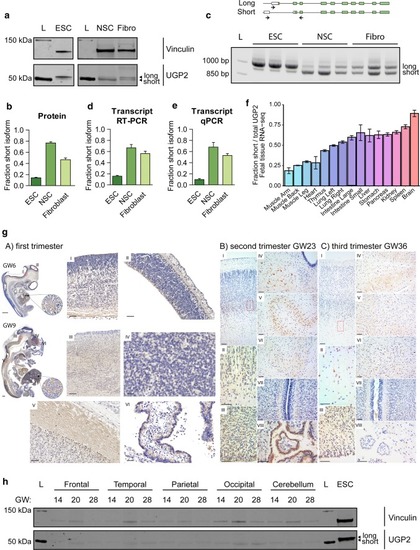
UGP2 short isoform is predominant in brain-related cell types. |
|
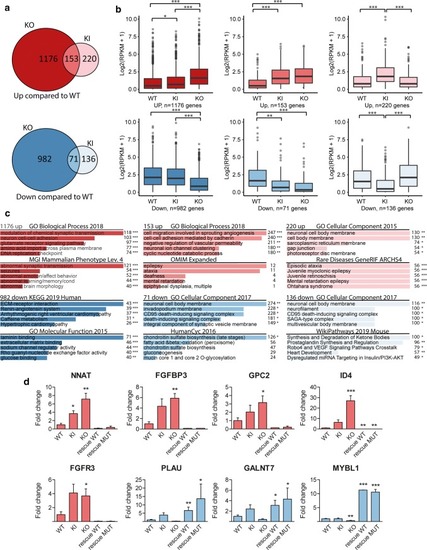
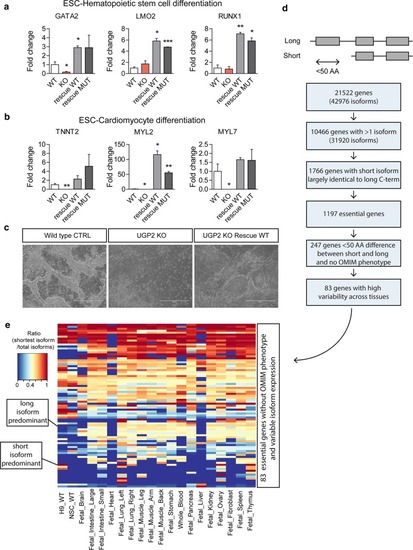
RNA-seq of UGP2 mutant H9-derived neural stem cells. |
|
Metabolic changes upon UGP2 loss. |
|
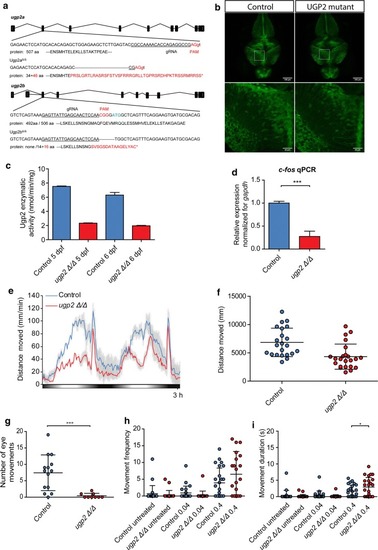
Zebrafish disease modeling. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Essentiality of UGP2 and other disease candidate genes with a similar mutation mechanism. |