- Title
-
A bi-modal function of Wnt signalling directs an FGF activity gradient to spatially regulate neuronal differentiation in the midbrain
- Authors
- Dyer, C., Blanc, E., Hanisch, A., Roehl, H., Otto, G.W., Yu, T., Basson, M.A., and Knight, R.
- Source
- Full text @ Development
|
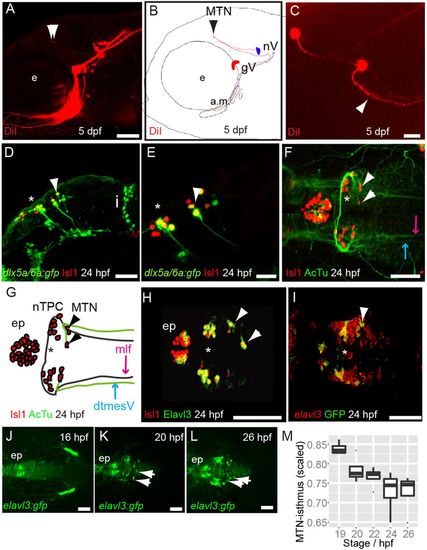
MTN neurons are the first differentiating neurons in the dorsal midbrain and innervate jaw muscles. Lateral view (A) and schematic (B) of 5 dpf zebrafish larvae with DiI (red) applied to the a.m. muscle showing labelled axons and MTN neurons (arrowheads) in the midbrain. Dorsal view (C) of DiI-labelled MTN neurons reveals a single axonal process that splits into two branches (arrowhead). Lateral views of 24 hpf Tg[dlx5a/6a:egfp] embryos with nTPC (asterisk) and MTN (arrowheads) labelled by GFP and Isl1 (D,E). Dorsal view and schematic of 24 hpf embryos with anti-Isl1 and acetylated tubulin (AcTu) labelling reveal that MTN neurons (arrowheads) pioneer the dtmesV (blue) and that nTPC neurons (asterisk) contribute to the mlf (purple) axon tracts (F,G). Dorsal views of 24 hpf embryos reveal that MTN and nTPC neurons are positive for Isl1 and Elavl3 (HuC) and express GFP in a Tg[elavl3:gfp] transgenic line (H,I); by contrast, more posteriorly located elavl3-expressing progenitor cells do not express GFP at similar stages (I). Dorsal views of Tg[elavl3:gfp] embryos at various stages reveal that MTN neurons (arrowheads) arise at the dorsal midline in the anterior midbrain and pioneer axon tracts ventrolaterally (J-L). A plot of the distance between MTN neurons and the isthmus, corrected for midbrain size, reveals that MTN neurons are formed at progressively posterior positions over time (M). gV, trigeminal ganglia; e, eye; a.m., adductor mandibulae; nV, trigeminal motoneurons; i, isthmus; mlf, medial longitudinal fasicle; ep, epiphysis; MTN, mesencephalic trigeminal nucleus; nTPC, nucleus of the tract of the posterior commissure; dtmesV, dorsal tract of the mesencephalic trigeminal. Scale bars: 100 μm in A,D,F-L; 20 μm in C,E. |
|
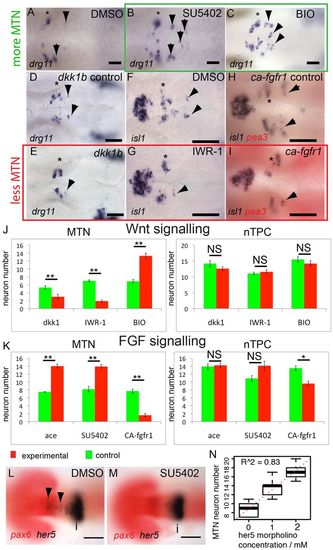
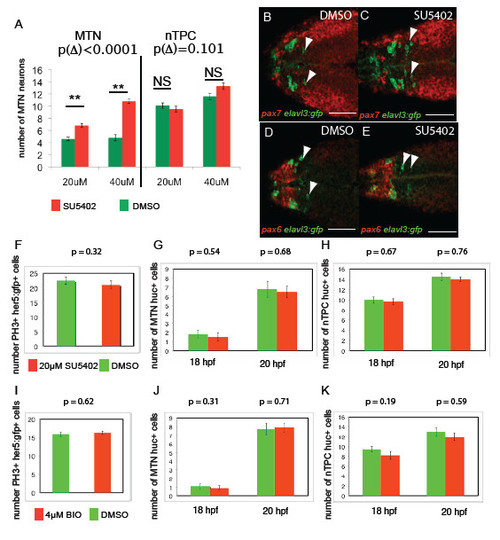
FGF, Wnt and Her5 dictate the number of MTN neurons that form in the midbrain. In situ hybridisation with probes for drg11 (A-E) and isl1 (F-I) reveals increased numbers of MTN neurons in zebrafish embryos exposed to 40 μM SU5402 (B) or 4 μM BIO (C) from 14 hpf (green box), relative to DMSO treatment (A). Overexpression of Dkk1b in Tg[hsp70l:dkk1b-egfp] embryos (D,E), treatment with 40 μM IWR-1 (F,G) or overexpression of a constitutively active Fgfr1 in Tg[hsp70:ca-fgfr1] embryos (H,I) also results in fewer MTN neurons (red box). Arrowheads indicate MTN neurons; asterisks indicate nTPC neurons. Quantification of neurons in animals with altered FGF (K) or Wnt (J) activity in the midbrain reveals a highly significant increase in MTN (P<0.01) but not nTPC (P>0.01) neuron number at 24 hpf for all conditions. ace, acerebellar. her5 expression in the dorsal midbrain (arrowheads) is lost by 20 hpf after application of 20 μM SU5402 at 14 hpf (M), but isthmus expression if unaffected relative to DMSO-treated controls (L). Plots of MTN and nTPC numbers in Her5 morphants reveals that loss of Her5 function causes a dose-dependent increase in MTN (R2=0.83) by 24 hpf (N). Error bars indicate s.e.m. **P<0.01; *P<0.05; NS, not significant; unpaired t-tests were used to compare between conditions; n=10 for each condition. i, isthmus. Scale bars: 100 μm in L,M; 20 μm in A-I. |
|
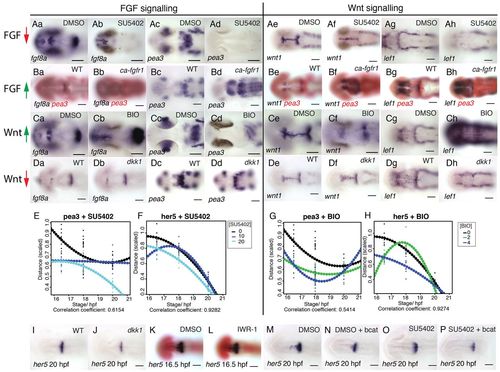
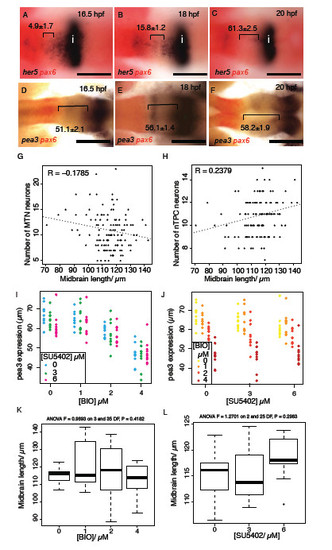
FGF and Wnt signalling interact to regulate each other’s activity. Dorsal views of zebrafish embryos processed for in situ hybridisation with probes to fgf8a (a,b in A-D), pea3 (c,d in A-D), wnt1 (e,f in A-D) and lef1 (g,h in A-D) following treatment with 40 μM SU5402 (Aa-h) or 4 μM BIO (Ca-h) or DMSO from 14 hpf or in transgenic Tg[hsp70:dkk1b-egfp] (Da-h), Tg[hsp70:ca-fgfr1] (Ba-h) and non-transgenic siblings after heat shock induction at 16.5 hpf. Spatial expression of pea3 (E,G) and her5 (F,H) across the dorsal midbrain was measured at 16.5, 18 and 20 hpf (n=10 for each stage) following exposure to SU5402 (E,F; 0, 10, 20 μM) or BIO (G,H; 0, 3, 6 μM). Polynomial plots (dotted line) were generated from quadratic equations describing the expression of each gene over time under the various conditions. Dorsal views of embryos processed by in situ hybridisation with probes to her5 (I-P) and pax6 (red, K,L) following treatment with 10 μM SU5402 (O,P), 40 μM IWR-1 (L) or DMSO from 14 hpf (K,M,N) in wild type (K,L), transgenic Tg[hsp70:dkk1b-egfp] (J) and non-transgenic siblings (I) or Tg[UAS:HA-bcat]; Tg[hsp70:gal4] (N,P) and non-transgenic siblings (M,O) after heat-shock induction at 16.5 hpf. Scale bars: 100 μm. |
|
Wnt regulation of Sprouty genes controls FGF activity at the isthmus and directs neuronal positioning. Zebrafish pea3, her5 and spry4 midbrain expression between 16.5 and 20 hpf (A); distance is scaled by midbrain size; error bars indicate s.e.m., n=30. Expression of spry4 at 24 hpf in embryos treated from 14 hpf with DMSO (B) or 4 μM BIO (C) or spry4 at 20 hpf in Tg[hsp70:dkk1b-egfp] transgenic embryos (E) and siblings (D) after heat-shock at 16.5 hpf. Expression (scaled to midbrain size) of spry4 in the dorsal midbrain between 16.5 and 20 hpf after exposure to SU5402 (F) or BIO (G) from 14 hpf. Quadratic polynomials (dotted line) were fitted to plots of spry4 expression (μm) over time after DMSO, SU5402 or BIO exposure from 14 hpf at varying concentrations (n=10 for each condition). Plots of pea3 and spry4 expression at 18 and 20 hpf, following heat-shock induction of dkk1 at 16.5 hpf in Tg[hsp70:dkk1b-egfp] embryos and non-transgenic siblings (H). Plots of spry4 expression at 24 hpf after exposure to varying SU5402 and BIO concentrations from 14 hpf were fitted with lines; the decreasing slopes of expression as BIO concentration is increased reveals that BIO upregulates spry4 and attenuates an SU5402-induced reduction of spry4 expression (I); data are average with s.e.m. and the model agreement is reported by the R2 value for linear models (n=90). Expression of Isl1 and Pax6 in E9.5 wild-type (J) and En1:Spry1/2-/- (K) mice was quantified (L) to reveal that MTN neurons are anteriorly displaced after loss of Sprouty function in the midbrain; values represent distance (µm) between the red arrow (MTN) and black arrow (isthmus); error bars indicate s.e.m. (n=10). Scale bars: 200 μm in J,K; 100 μm in D,E; 50 μm in B,C. |
|
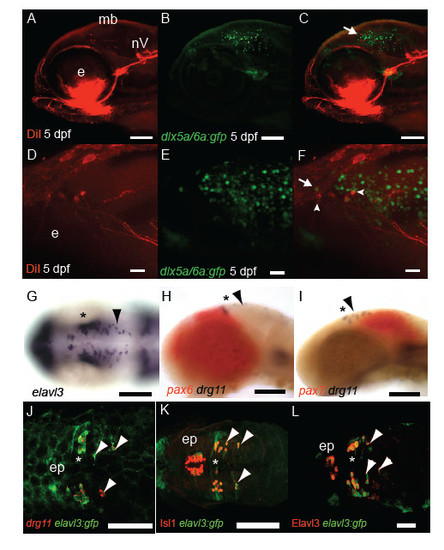
Characterisation of MTN neuronal development. DiI labelling (A,D) of jaw muscles and GFP (B,E) in Tg[dlx5a/6a:egfp] 5 dpf larvae reveals that a proportion of MTN neurons (arrowheads) co-express GFP and are larger than adjacent tectal GFP+ neurons (arrow, C,F). At 24 hpf many huc /elavl3 expressing progenitor cells are present along the A-P extent of the midbrain (G). In contrast drg11 expressing MTN (arrowheads) are restricted to the pax7 expressing midbrain (I) in comparison to nTPC neurons present in the pax6+ forebrain (H). In Tg[elavl3:gfp] embryos at 24 hpf MTN and nTPC neurons are GFP+ and express drg11 (J), Isl1 (K) and HuC protein (L). Scale bars 100 μm (A-C,G-K) 50μm (L) 20μm (D-F). |
|
FGF and Wnt signalling regulates MTN number between 14-24 hpf, but not nTPC number, progenitor cell specification or proliferation. Treatment of embryos from 14 hpf with SU5402 at 20μM compared to 40μM results in a statistically significant increase in number (A, difference of change Δ following SU5402 treatment compared to DMSO, p Δ = 9.51x10-5, n=30). Transgenic Tg[elavl3:gfp] embryos exposed to 40μM SU5402 from 14 hpf (C) reveal that ectopic MTN neurons (green, anti-GFP) form within the pax7 expressing midbrain (red, pax7 in situ) at 24 hpf relative to DMSO treated controls (F), but are not forming in pax6+ forebrain regions (D,E). Quantification of proliferating cells in the midbrain of Tg[her5:gfp] embryos at 18 hpf following exposure to 20μM SU5402 (F) or 4μM BIO (I) from 14 hpf reveals no significant differences relative to DMSO treated control animals (p>0.05 for all, n=20). Quantification of huc/ elavl3 expressing neuronal progenitors in the midbrain or posterior diencephalon of 20 hpf and 18 hpf embryos treated with 20μM SU5402 (G,H) or 4μM BIO (J,K) from 14 hpf reveals no significant differences relative to DMSO treated control animals (p >0.05 for all, n=20). Error bars represent s.e.m., statistical comparisons were performed using unpaired t-tests. Scale bars 100μm 50μm (B-E). |
|
FGF-responsive genes show a posterior shift during midbrain development and FGF and Wnt signalling interact to regulate pea3 expression during midbrain development. Dorsal views of embryos processed by in situ reveals her5 (A-C) and pea3 (D-F) expression relative to pax6 (forebrain); expression retracts posteriorly towards the isthmus over time (distance between midbrain expression boundary – pax6 in μm, ± s.e.m., n=10). Plots of midbrain size relative to MTN number (G) and nTPC number (H) from animals treated with varying doses of SU5402 or BIO reveals no correlation (n=240). Plots of pea3 expression at 24 hpf after application of varying SU5402 (I) or BIO (J) doses (on x-axis) reveal that BIO application results in a strong downregulation of pea3 independently from the reduction of expression caused by SU5402 (dots represent individual samples, n=120). Plots of midbrain length in 24 hpf embryos treated with BIO (0, 1, 2, 4μM) or SU5402 (0, 3, 6μM) from 14 hpf (K,L). ANOVA tests for significance did not show any linkage between BIO (p=0.42) or SU5402 (p=0.30) concentration and midbrain length. Isthmus (i). Scale bars 100μm (A-F). |
|
Wnt and FGF signalling regulate each others activity through feedback loops. Dorsal views of embryos processed by in situ hybridisation with probes to dgfp (A-E), fgf8a (F-I), pea3 (J-M), wnt1 (N-Q), lef1 (R-U) in 24 hpf embryos from Tg[top:dgfp] (A-E) or Tg[UAS:HA-bcat]; Tg[hsp70:gal4] (F-U) transgenics. Embryos were treated with 10μM SU5402 (H,I,L,M,P,Q,T,U) or DMSO (F,G,J,K,N,O,R,S) from 14 hpf, then heat-shocked at 16.5 hpf; embryos over-expressing betacatenin were sorted based on HA expression. Scale bars 100μm (A-U). |
|
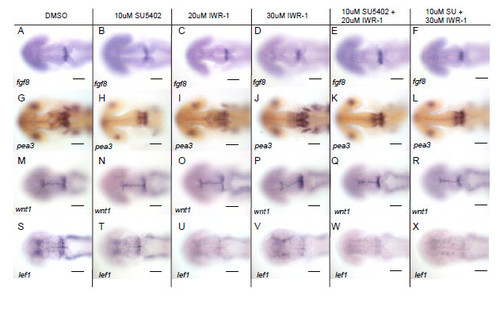
Wnt signalling does not directly regulate FGF activated genes. Dorsal views of embryos processed by in situ hybridisation with probes to fgf8a (A-F), pea3 (G-L), wnt1 (M-R), lef1 (S-X) in 24 hpf embryos following treatment with DMSO, 10μM SU5402, 20μM or 30μM IWR-1 or combinations of 10μM SU5402 with 20μM or 30μM IWR-1 from 14 hpf. Scale bars 100μm. |
|
Wnt signalling regulates FGF activity in the midbrain through Sprouty genes. Results from microarray analyses of gene expression showing fold change after upregulation of Wnt8b (column 1) or Dkk1b (column 3) expression (A). P value indicates significance of gene expression changes relative to global changes in gene expression and are colour coded to reflect the fold change. Overall responses to changes in Wnt signalling (genes showing >1.5-fold response to Wnt or Dkk1 upregulation) are represented in column 5 with values >1 representing a consistent response by the gene to changes to Wnt. Dorsal views of spry4 (blue) and pax6 (red) expression in embryos at 20 hpf (values represent distance in μm, ± s.e.m.) treated from 14 hpf with DMSO (B), 20μM SU5402 (C), 4μM BIO (D), 2μM BIO and 10μM SU5402 (E), 4μM BIO and 20μM SU5402 (F). Plots of spry4 expression at 20 hpf after application of varying SU5402 and BIO doses reveals that upregulated Wnt signalling can cause upregulation of spry4 even in the presence of SU5402 (G, H; dots represent individual samples, n=90). Dorsal views of Tg[UAS:HA-bcat]; Tg[hsp70:gal4] embryos processed by in situ hybridisation to examine spry4 expression at 24 hpf after heat-shock at 16.5 hpf and addition of DMSO or 10μM SU5402 (I-L). Scale bars 100μm (I-L), 50μm (A-F). |