- Title
-
Igf Signaling is Required for Cardiomyocyte Proliferation during Zebrafish Heart Development and Regeneration
- Authors
- Huang, Y., Harrison, M.R., Osorio, A., Kim, J., Baugh, A., Duan, C., Sucov, H.M., and Lien, C.L.
- Source
- Full text @ PLoS One
|
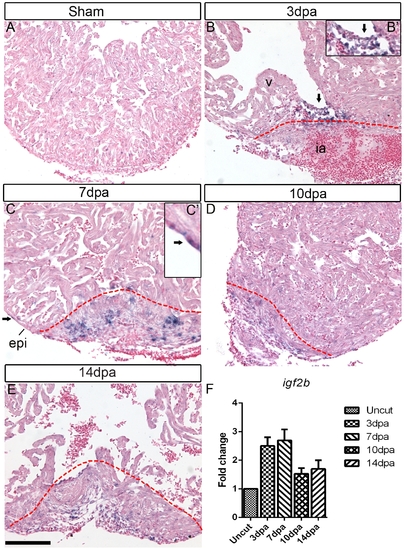
igf2b is upregulated during zebrafish heart regeneration. In situ hybridization (ISH) was performed on sham operated hearts (A) and 3 dpa (B), 7 dpa (C), 10 dpa (D) and 14 dpa (E) regenerating hearts. There is no detectable igf2b expression in sham-operated hearts by ISH, whereas strong expression is induced in endocardium (arrows in B and inset B′) and epicardium (arrows in C and inset C′) near the wound following amputation of the ventricular apex. Red dashed lines mark the amputation site and wound area. v: ventricle, ia: injured area, epi: epicardium. Scale bar = 100 μm. (F) Quantitative RT-PCR of igf 2b expression at 3, 7, 10 and 14 dpa compared to uncut hearts. |
|
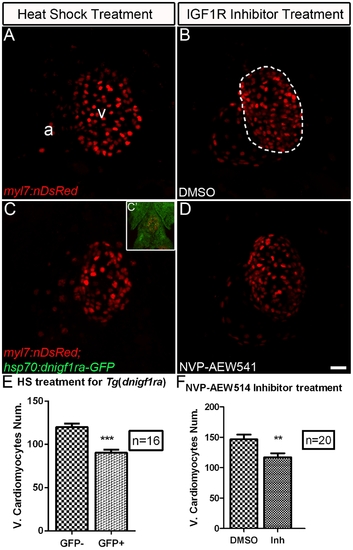
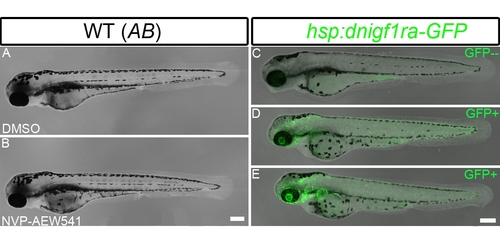
Igf signaling is required for proper cardiomyocyte number in zebrafish embryonic hearts. The number of ventricular cardiomyocytes (area encircled by dotted line) number decreased in embryos when Igf1r signaling was blocked. (A) Tg(myl7:nDsRed) single transgenic and (C) Tg(hsp:dnigf1ra-GFP; myl7:nDsRed) double transgenic embryos (n = 16) were heat shocked at 40°C for 30 min around 48 and 72 hpf and observed at 76 hpf. dnigf1ra embryos can be identified by GFP expression (C’ inset). Tg(myl7:nDsRed) single transgenic siblings (n = 16) were used as controls. myl7:nDsRed embryos were treated with Igf1r inhibitor (D) (n = 20) and DMSO as a control (B) (n = 20) from 48 to 72 hpf and observed at 76 hpf. a: atrium, v: ventricle. Scale bar = 20 μm. (E, F) Quantification of mean ventricular cardiomyocyte number ± S.E. (***p<0.0001; **p<0.001). Fewer ventricular cardiomyocytes were observed around 76 hpf after Igf1r signaling was blocked. |
|
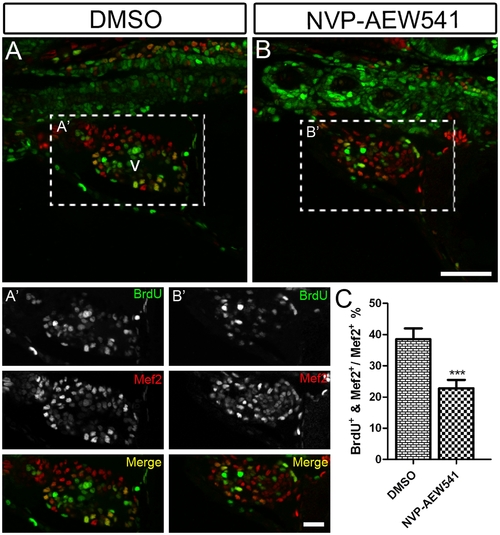
Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development. Wild type fish were treated with DMSO as a control (n = 5) (A and A′) and the Igf1r inhibitor NVP-AEW541 (n = 7) (B and B′) from 48–72 hpf. BrdU was added for the same time period. BrdU (green) and Mef2 (red) double positive cells indicate proliferating cardiomyocytes (A, A′, B, B′). A′ and B′ are images of the dashed boxes in A and B. (A′ and B′), BrdU (green) and Mef2 (red) staining were shown as black and white or merged color images. Scale bar: (B) = 50 μm, (B′) = 20 μm. v: ventricle. (C) Quantification of BrdU positive cardiomyocytes (Mef2 positive) ± S. E. A significant decrease (***p<0.0001) in cardiomyocyte proliferation was detected in embryos treated with NVP-AEW541. |
|
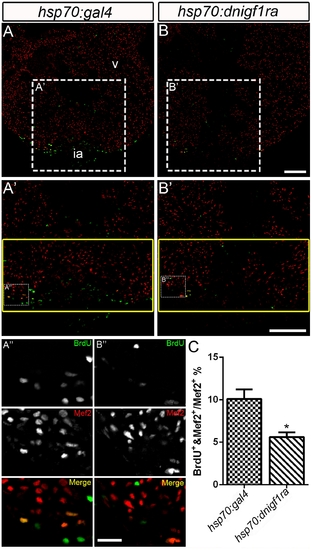
Igf signaling is required for cardiomyocyte proliferation during zebrafish heart regeneration. Cardiomyocyte proliferation was decreased after Igf signaling was blocked during zebrafish heart regeneration. Tg(hsp70:gal4) control (n = 5) (A, A′, A′′) and Tg(hsp70:dnigf1ra-GFP) transgenic zebrafish (n = 7) (B, B′,B′′) zebrafish were heat shocked for 1 h at 38°C after amputation from 2–10 dpa. BrdU (green) and Mef2 (red) double positive cells indicate proliferating cardiomyocytes (A, A′, B, B′). A′ and B′ are the higher magnification images of the dashed boxes in A and B. A′′ and B′′ are the higher magnification images of the dashed boxes in A′ and B′. The yellow box indicates the wound area; cardiomyocytes were counted in this region. (A′′ and B′′), BrdU (green) and Mef2 (red) staining were shown as separated channel images (black and white) or merged color images. ia: injured area, v: ventricle. Scale Bar: (B, B′) = 100 μm, (B′′) = 20 μm. (C) Quantification of BrdU positive cardiomyocytes (Mef2 positive) ± S.E. A significant decrease (*p<0.05) in cardiomyocyte proliferation was detected in Tg(hsp:dnigf1ra-GFP) fish. |
|
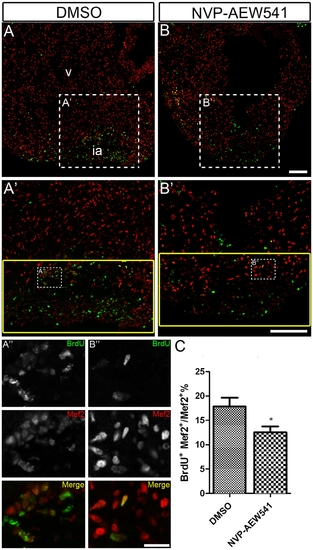
Chemical inhibition of Igf signaling suppresses cardiomyocyte proliferation during zebrafish heart regeneration. Wild type fish were treated with DMSO as a control (A, A′ and A′′) (n = 5) and the Igf1r inhibitor NVP-AEW541 (B, B′ and B′′) (n = 5) from 2-14 dpa. BrdU (green) and Mef2 (red) double positive cells indicate proliferating cardiomyocytes (A, A′, B, B′). A′ and B′ are the higher magnification images of the dashed boxes in A and B. A′′ and B′′ are the higher magnification images of the dashed boxes in A′ and B′. The yellow box indicates the wound area and cardiomyocytes were counted in this region. (A′′ and B′′), BrdU (green) and Mef2 (red) staining were shown as black and white or merged color images. Scale bar: (B, B′) = 100 μm, (B′′) = 20 μm. ia: injured area, v: ventricle. (C) Quantification of BrdU positive cardiomyocytes (Mef2 positive) ± S.E. A significant decrease (*p<0.01) in cardiomyocyte proliferation was detected in fish treated with Igf1r inhibitor NVP-AEW541. |
|
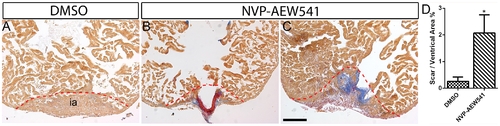
Igf signaling is required for heart regeneration. Wild type fish were treated DMSO (A) (n = 6) or the Igf1r inhibitor NVP-AEW541 (n = 7) (B and C) from 2–30 dpa. AFOG staining was performed at 30 dpa to detect collagen (blue) and fibrin (red) deposition. The dashed line marks the regenerating area. Scale bar = 100 μm. ia: injured area. (D) Quantification of scar area normalized to ventricle area in DMSO and NVP-AEW541 treated hearts. A significant increase (*p<0.05) in scar/ventricular area ratio was detected in NVP-AEW541 treated hearts. |
|
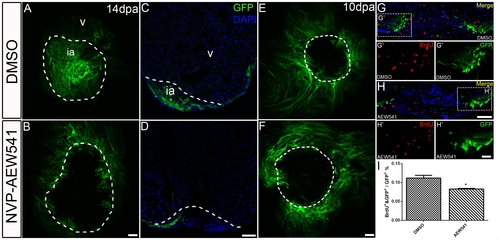
Igf signaling is required for contribution ofgata4:EGFP positive cardiomyocytes to the regenerating area. gata4:EGFP fish were treated with the Igf1r inhibitor NVP-AEW541 from 2-14 dpa (n = 12) (B, D) and 7-10 dpa (n = 5) (F, H). DMSO was used as a control (14dpa: n = 10; 10 dpa: n = 6) (A, C, E, G). Whole mount confocal microscopy from the view of the apex (A, B, E, F) and frozen sections (C, D, G, H) were performed at 14 and 10 dpa to determine the contribution of the EGFP positive population. BrdU (red) and Gata4 (green) double positive cells indicate proliferating gata4:EGFP positive cardiomyocytes (G, G′, H, H′). G′, and H′ are the higher magnification images of the dashed boxes in G and H. BrdU staining (red) and gata4:EGFP (green) were shown as separated channel images. The dashed line marks the regenerating area. Scale bar: (B, D, F, H) = 50 μm; (H′) = 20 μm. ia: injured area, v: ventricle. (I) Quantification of BrdU and gata4:EGFP double positive cells/gata4:EGFP area ± S.E. A significant decrease (*p<0.05) in gata4:EGFP positive cell proliferation was detected in fish treated with Igf1r inhibitor NVP-AEW541 from 7-10dpa. |
|
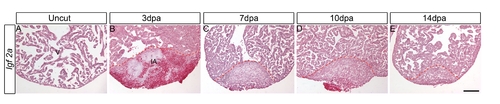
igf2a expression is not detected during zebrafish heart regeneration. ISH was performed on uncut hearts (A) and 3 dpa (B), 7 dpa (C), 10 dpa (D) and 14 dpa (E) regenerating hearts. There is no detectable igf2a expression in regenerating hearts by ISH. Scale bar = 100 μm. v: ventricle; ia: injured area. It was shown previously that embryos injected with igf1r morpholinos exhibited severely reduced body length at 24 hpf [43], raising the possibility that blocking Igf signaling inhibits overall embryonic growth and development in general. We examined embryo length and overall development of Tg(hsp70:dnigf1ra-GFP) embryos or embryos treated with the Igf1r chemical inhibitor NVP-AEW-541 from 48–72 hpf. We observed only a very mild reduction in the length of the inhibitor treated (3.7%) and transgenic embryos (5.7%) (Figure S2. A-E). These results suggest that the reduced cardiomyocyte numbers are unlikely caused by effects of Igf signaling on overall embryo growth and development indicating Igf signaling is required for zebrafish heart development. |
|
Inhibiting Igf signaling in zebrafish embryos results in mild defects in overall length. WT embryos were treated with DMSO as a control (A) (n = 20) or the Igf inhibitor NVP-AEW541 (B) (n = 20) from 48–72 hpf. Tg(hsp70:dnigf1ra-GFP) (D and E) and non-transgenic control (C) embryos were heat shocked twice at 48 and 72 hpf for 30 mins at 40°C. The embryos were observed at 76 hpf using a confocal microscope. Tg(hsp70:dnigf1ra-GFP) were identified by GFP expression. The length of the inhibitor treated embryos is about 3.2% shorter than untreated control embryos, and the Tg(hsp70:dnigf1ra-GFP) transgenic embryos are 5.7% % shorter than non-transgenic control embryos. Scale bar: (B) = 200 μm. |
|
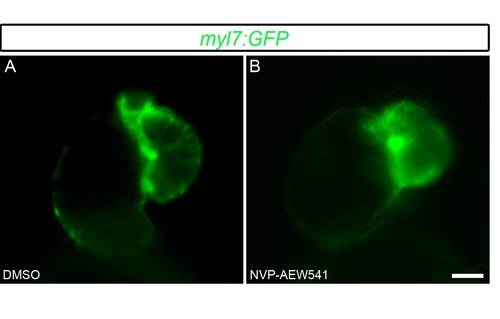
Inhibiting Igf signaling in zebrafish embryos results in abnormal cardiac looping. Tg(myl7:GFP) embryos were treated with DMSO as a control (A) (n = 20) or the Igf inhibitor NVP-AEW541 (B) (n = 20) from 48–72 hpf. The embryos were observed at 76 hpf using a fluorescence microscope. Scale bar = 20 μm. |
|
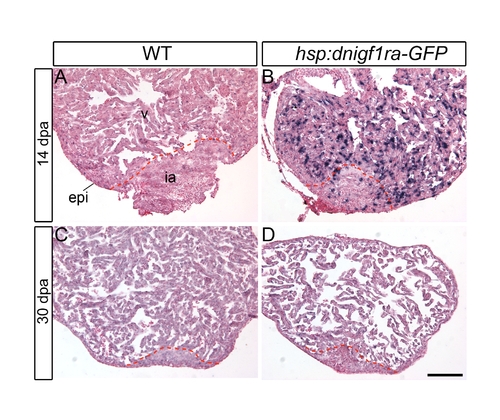
Down regulation of the dnigf1ra transgene is detected after 30 days of heat shock. Wild type or Tg(hsp70:dnigf1ra-GFP) were heat shocked from 2–14 or 2–30 dpa. ISH was performed to assess dnigf1ra transgene expression in 14 dpa and 30 dpa regenerating hearts. Strong dnigf1ra transgene expression was detected at 14 dpa (B). Weak or no expression of the dnigf1ra transgene was detected at 30 dpa (D). Scale bar = 100 μm. v: ventricle, ia: injured area, epi: epicardium. |
|
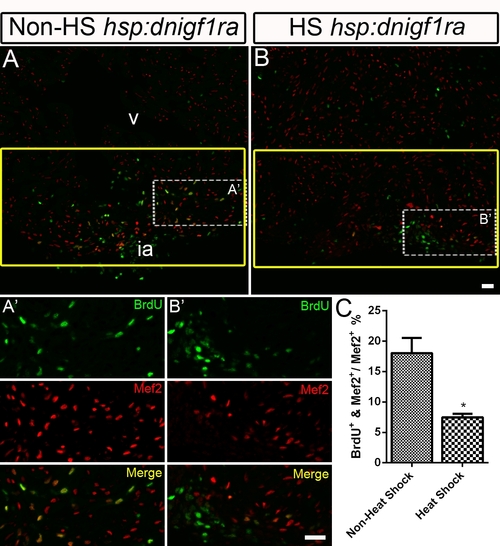
Non heat-shocked control for the Tg(hsp70:dnigf1ra-GFP) fish experiment. BrdU incorporation was determined in non heat-shocked (Non-HS) Tg(hsp70:dnigf1ra-GFP) control fish (n = 4) (A and A′) and heat shocked (HS) Tg(hsp70:dnigf1ra-GFP) transgenic zebrafish (n = 6) (B and B′). Tg(hsp70:dnigf1ra-GFP) transgenic were heat shocked for 1 h at 38°C after amputation from 2–10 dpa. Non heat-shocked control fish were kept in the regular system. BrdU (green) and Mef2 (red) double positive cells indicate proliferating cardiomyocytes (A, A′, B, B′). A′ and B′ are the higher magnification images of the dashed boxes in A and B. The yellow box indicates the wound area; cardiomyocytes were counted in this region. BrdU (green) and Mef2 (red) staining were shown single channeled or merged color images. ia: injured area, v: ventricle. Scale Bar = 20 μm. (C) Quantification of BrdU positive cardiomyocytes (Mef2 positive) ± S.E. A significant decrease (*p<0.05) in cardiomyocyte proliferation was detected in Tg(hsp70:dnigf1ra-GFP) fish. |