- Title
-
Dhx34 and Nbas function in the NMD pathway and are required for embryonic development in zebrafish
- Authors
- Anastasaki, C., Longman, D., Capper, A., Patton, E.E., and Cáceres, J.F.
- Source
- Full text @ Nucleic Acids Res.
|
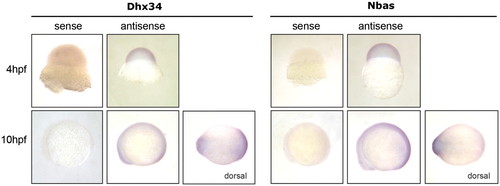
Dhx34 and Nbas are ubiquitously expressed during zebrafish embryogenesis. Wholemount in situ hybridization against dhx34 and nbas using antisense riboprobes revealed ubiquitous expression of dhx34 and nbas transcripts (seen as purple staining) during early zebrafish development. No significant staining was detected using the control sense probes. |
|
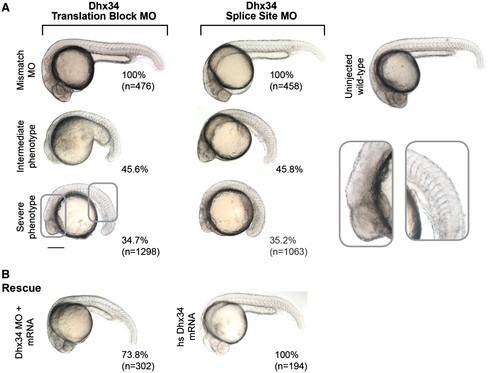
Dhx34 is required for proper embryonic development in zebrafish. (A) Translation block and splice site MOs against zebrafish Dhx34 cause a distinctive range of developmental phenotypes at 24hpf characterized by shortening of the anteroposterior axis, somite malformation and significant reduction in head size. Morphants injected with the mismatch control MOs are indistinguishable from uninjected wild-type embryos. (B) Co-injection of Dhx34 translation block MO and human full-length DHX34 mRNA rescued the morphant phenotype in 73.8% of the injected embryos at 24 hpf. Expression of the mRNA alone has no overt effect on development. The numbers beside each panel indicate the percentage of embryos with the observed phenotype. Numbers in brackets correspond to the total of analysed embryos. PHENOTYPE:
|
|
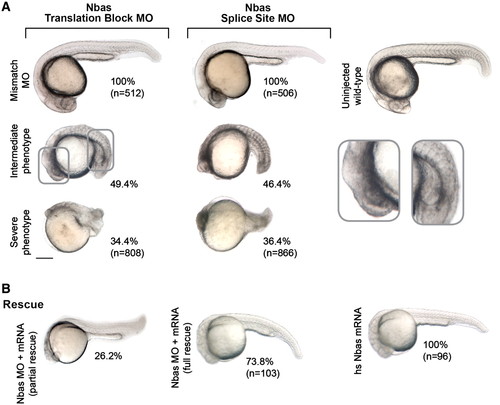
Nbas is essential for zebrafish embryonic development. (A) Injections with translation block and splice site MOs against zebrafish nbas reproduced the developmental phenotype observed in Upf1 and Dhx34 morphants. (B) Co-injection of Nbas translation block MO and human full length NAG/Nbas mRNA rescued the morphant phenotype in the majority of the embryos at 24 hpf. Expression of the mRNA alone had no effect on embryonic development. The numbers beside each panel represent the percentage of embryos showing each phenotype and in brackets is the total number of the analysed embryos. PHENOTYPE:
|
|
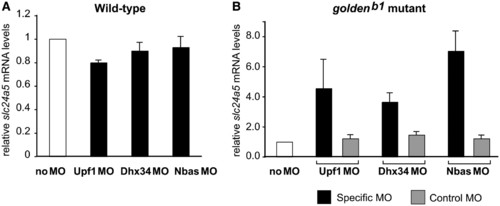
Both Dhx34 and Nbas are required for the in vivo degradation of the nonsense slc25a5 transcript in zebrafish. Expression of slc25a5 mRNA was measured in wild-type (A) and goldenb1 mutant (B) uninjected embryos, or embryos injected with indicated MOs, by real-time RT-PCR. The level of slc25a5 transcript was normalized to dct mRNA. The normalized values were then divided by values for uninjected samples (no MO) and expressed as relative mRNA levels. EXPRESSION / LABELING:
|
|
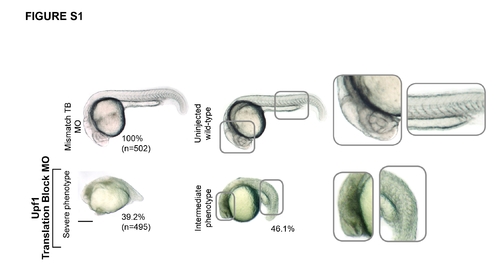
Upf1 is required for proper embryonic development in zebrafish. (A) Translation block MOs against zebrafish Upf1 caused severe developmental delay phenotypes at 24hpf characterized by shortening of the anteroposterior axis, somite malformation and brain necrosis indicated by opaque brain tissue. Morphants injected with the mismatch control MOs show no developmental defects similar to uninjected wild-type embryos. The numbers beside each panel indicate the percentage of embryos with the displayed phenotype, whereas numbers in brackets correspond to the total of embryos analyzed. PHENOTYPE:
|