- Title
-
Semaphorin signaling guides cranial neural crest cell migration in zebrafish
- Authors
- Yu, H.H., and Moens, C.B.
- Source
- Full text @ Dev. Biol.
|
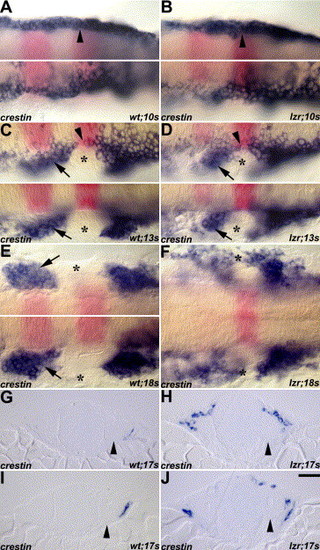
Migrating cranial NCCs in wild-type (A, C, E, G, I) and lzr/pbx4 mutant (B, D, F, H, J) embryos revealed by crestin RNA in situ hybridization (blue). Flat-mount embryos are arranged with anterior to the left and krox20 (pink) indicates r3 and r5. Arrows indicate the second cranial NC stream in C–E. (A–B) Cranial NCCs (arrowheads) were observed as bilateral homogenous populations at the 10s stage both in the wild-type and lzr/pbx4 mutant embryos. (C–D) The separated cranial NCC streams and the cranial NC-free zone (asterisks) were observed at 13s stage both in wild-type and lzr/pbx4 mutant embryos. (E–F) The fusion of cranial NC streams in the periphery was observed at 18s stage in lzr/pbx4 mutants (asterisks in F), while the cranial NC-free zone was observed at the same stage of wild-type embryos (asterisk in E). The top and bottom panels in A, B, and E are different focal planes on both sides of the same embryo. (G–J) The distribution of cranial NCCs around the anterior OV (arrowheads) is shown in the two sequential cross sections of both the wild-type and lzr/pbx4 mutant embryos. G and H are the cross sections anterior to I and J, respectively. Scale bar: 20 µm in A–J. EXPRESSION / LABELING:
PHENOTYPE:
|
|
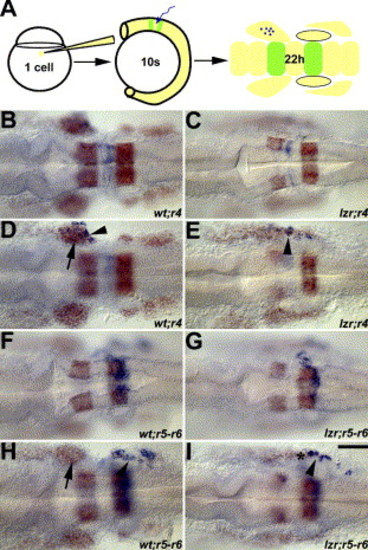
Mis-migrated lzr/pbx4 cranial NCCs are derived from r4. (A) Schematic figure of the uncaging experiment for tracing the migration of cranial NCCs. The cells labeled by laser uncaging are shown in blue and krox20 and dlx2 (for cranial NCCs) are shown in orange. B, C, F, and G are dorsal images of D, E, H, and I, respectively, to show the labeled rhombomeres. In wild-type embryos (B, D, F, H), labeled r4 and r5–r6 cranial NCCs (arrowhead) migrate into the second and third cranial NC streams, respectively (D and H). In lzr/pbx4 mutants (C, E, G, I), cells marked in r4 but not in r5–r6 give rise to cranial NCCs in the normally cranial NC-free zone. Asterisks mark the region with the mis-migrated lzr/pbx4 cranial NCCs in E and I. Scale bar: 40 μm in B–I. |
|
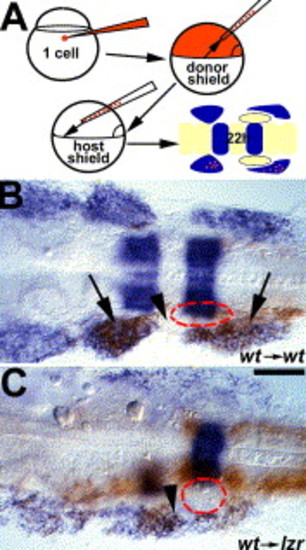
lzr/pbx4 functions non-cell autonomously to control cranial NCC migration. (A) Schematic of the approach for generating cranial NCC mosaics. Donor-derived cells are brown, dlx2 and krox20 expression are in blue. Red dash-lines circle the position of the OV in B-C. (B) In control experiments, wild-type donor cells migrate normally into the second and third cranial NC streams of the pharyngeal arches (arrows) without occupying the cranial NC-free zone (arrowhead) in a wild-type host embryo. (C) In contrast, wild-type donor cells are seen in the normally cranial NC-free zone (arrowhead) after transplantation into a lzr/pbx4 mutant host. Scale bar: 40 μm in B–C. |
|
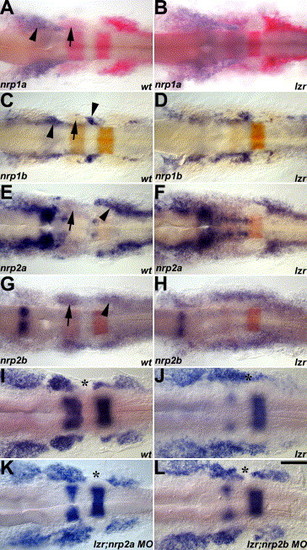
Zebrafish nrps interact with lzr/pbx4 during cranial NCC migration. nrp expression is shown in blue in A–H; krox20 and dlx2 are in pink or orange in A–H and blue in I–L. Arrows indicate the second cranial NC stream. (A–B) nrp1a is expressed on the first cranial NC stream (arrowhead). (C–D) nrp1b is expressed in a subset of proximal cranial NCCs contributing to the cranial sensory ganglia (arrowheads). (E–H) nrp2a in E-F and nrp2b in G–H are expressed in cranial NCCs. nrp2a is expressed strongly on the first and third cranial NC streams and weakly on the second cranial NC stream, while nrp2b is expressed in all three streams. Arrowheads indicate the third cranial NC stream in E and G. In lzr/pbx4 mutants, nrp expression in the cranial NC is disorganized in a manner corresponding to the disorganization of cranial NCCs. (I–L) MO knockdown of Nrp2a (K) or Nrp2b (L) results in the restoration of the normal cranial NCC migratory pattern. Asterisks mark the cranial NC-free zone in I. K and L and cranial NCCs in this normally cranial NC-free zone in lzr/pbx4 mutants (J). Scale bar: 40 μm in A–L. EXPRESSION / LABELING:
PHENOTYPE:
|
|
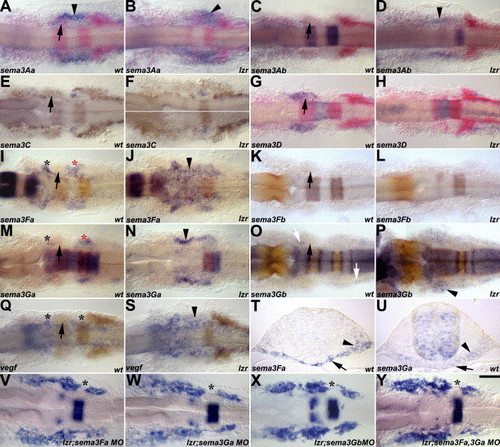
Class 3 secreted semas are expressed in the cranial NC-free zones. In A–U, sema expression is in blue, and engrailed3, krox20, and dlx2 are in pink or orange. In V–Y, krox20 and dlx2 are in blue. Arrows indicate the second cranial NC stream. (A–B) sema3Aa is expressed in the head periphery between the second and third cranial NC streams (arrowhead), and its expression appears unaffected in lzr/pbx4 mutants. (C–D) sema3Ab is not expressed in the head periphery of wild-type embryos, but its expression is expanded in the head periphery between the second and third cranial NC streams of lzr/pbx4 mutants (arrowhead). (E–F) sema3C is expressed in the OV (arrowhead) in wild-type embryos, and its expression is also unaffected in lzr/pbx4 mutants (G–H) sema3D is expressed in the second cranial NC stream (arrow) in wild-type embryos, but the expression is gone in lzr/pbx4 mutants. sema3Fa (I–J), sema3Ga (M–N), and vegf165 (Q–S) are expressed in the cranial NC-free zones (asterisks) in wild-type embryos and their expression is expanded in the head periphery of the lzr/pbx4 mutants (arrowhead). (K–L) sema3Fb is not expressed in the head periphery. (O–P) sema3Gb is expressed in cells surrounding the cranial NCCs in the wild-type embryo (white arrows) and its expression is expanded in the head periphery of lzr/pbx4 mutants (arrowhead). (T–U) The expression of sema3Fa (T) and sema3Ga (U) in the endoderm (arrows) and/or mesoderm (arrowheads) of the cranial NC-free zones in wild-type embryos is examined in cross sections, whose positions are indicated as red asterisks in I and M. (V–Y) MO knockdown of Sema3Fa (V), Sema3Ga (W), Sema3Gb (X), or Sema3Fa/3Ga (Y) in lzr/pbx4 mutants partially suppresses the cranial neural crest fusion phenotype, resulting in the re-appearance of a cranial NC-free zone (asterisks) in V–Y. Scale bar: 40 μm in A–S and V–Y; 20 μm in T–U. EXPRESSION / LABELING:
PHENOTYPE:
|
|
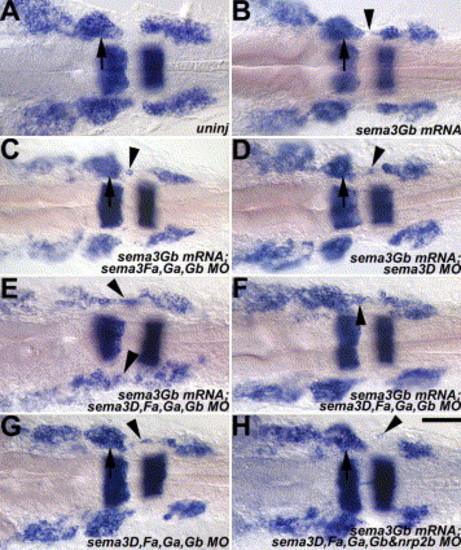
Over-expression of sema3Gb in wild-type embryos results in aberrant cranial NCC migration. Manipulation in each panel is under the wild-type genetic background. krox20 and dlx2 are in blue in all panels. Arrows indicate the second cranial NC stream, while arrowheads indicate cranial NCCs in normally cranial NC-free zones. (A) An uninjection control of a wild-type embryo. (B) A mild cranial NCC migration defect (arrowhead), in which a few cranial NCCs were detected in the normally cranial NC-free zone, was occasionally observed in sema3Gb over-expressing embryos. (C) Over-expression of sema3Gb combined with knockdown of endogenous Sema3Fa, Sema3Ga, and Sema3Gb causes a similar mild defect. (D) Knockdown of Sema3D in the presence of over-expression of sema3Gb had a mild defect. (E-F) Over-expression of sema3Gb in wild-type embryos injected with Sema3D, Sema3Fa, Sema3Ga, and Sema3Gb MOs caused a cranial NCC migration phenotype resembling that of lzr/pbx4 mutants. One (arrowhead in F) or both (arrowheads in E) sides of the manipulated embryos showed the cranial NC fusion phenotype. (G) Knockdown of endogenous Sema3D, Sema3Fa, Sema3Ga, and Sema3Gb also had a similar mild defect. (H) Knockdown of Nrp2b suppresses the cranial NC fusion phenotype observed in the manipulated embryos of E and F. An aberrantly migratory cranial NCC is indicated by arrowhead. Scale bar: 40 μm in A–H. |
Reprinted from Developmental Biology, 280(2), Yu, H.H., and Moens, C.B., Semaphorin signaling guides cranial neural crest cell migration in zebrafish, 373-385, Copyright (2005) with permission from Elsevier. Full text @ Dev. Biol.