- Title
-
Modulating the Inflammatory Response to Wounds and Cancer Through Infection
- Authors
- López-Cuevas, P., Cross, S.J., Martin, P.
- Source
- Full text @ Front Cell Dev Biol
|
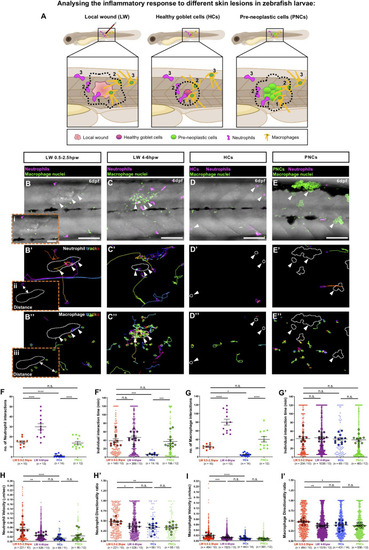
Analysing the inflammatory response to different skin lesions in zebrafish larvae. |
|
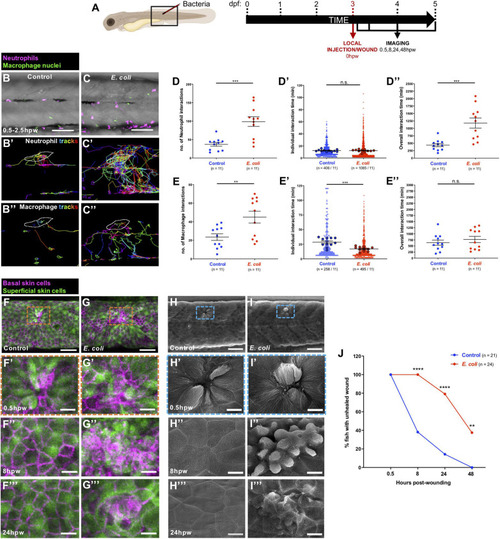
Alterations to the wound inflammatory response upon |
|
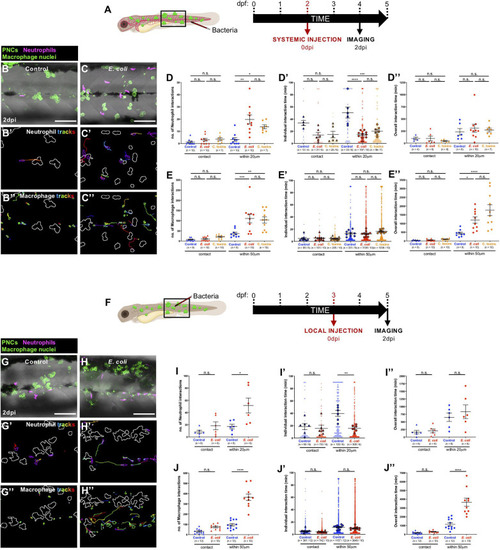
Altered cancer inflammatory response upon |
|
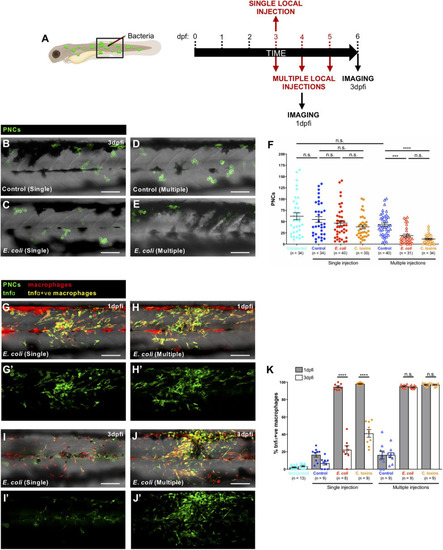
Prolonged pro-inflammatory response and reduction in cancer cell numbers upon consecutive local injections of |