- Title
-
Inappropriate cathepsin K secretion promotes its enzymatic activation driving heart and valve malformation
- Authors
- Lu, P.N., Moreland, T., Christian, C.J., Lund, T.C., Steet, R.A., Flanagan-Steet, H.
- Source
- Full text @ JCI Insight
|
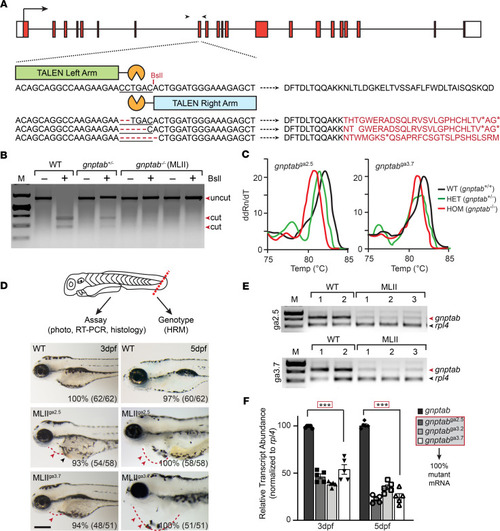
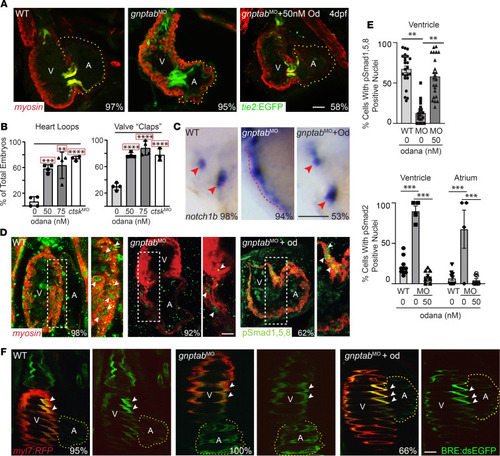
( EXPRESSION / LABELING:
PHENOTYPE:
|
|
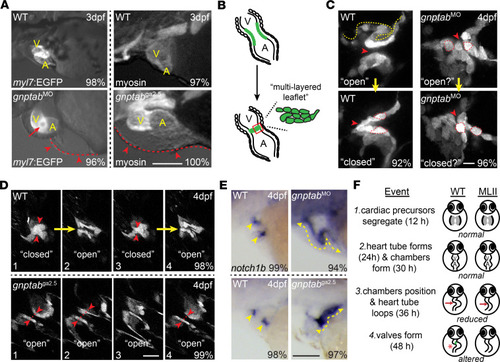
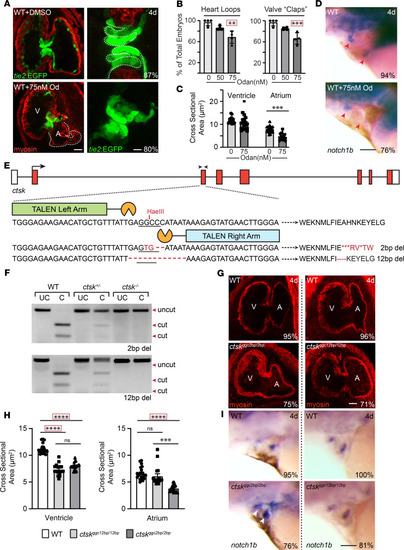
( EXPRESSION / LABELING:
PHENOTYPE:
|
|
( |
|
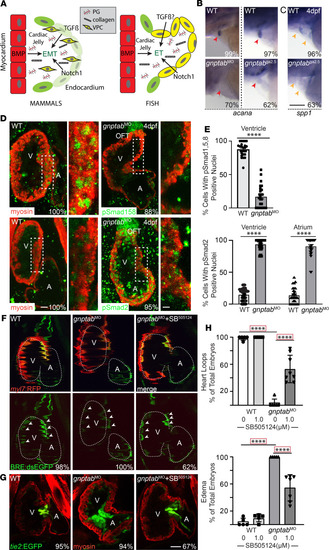
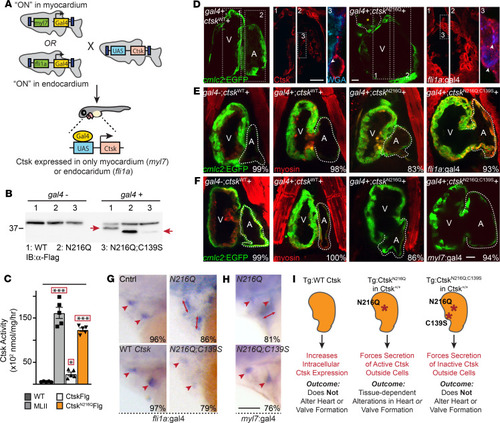
( EXPRESSION / LABELING:
|
|
( |
|
( |
|
( |