- Title
-
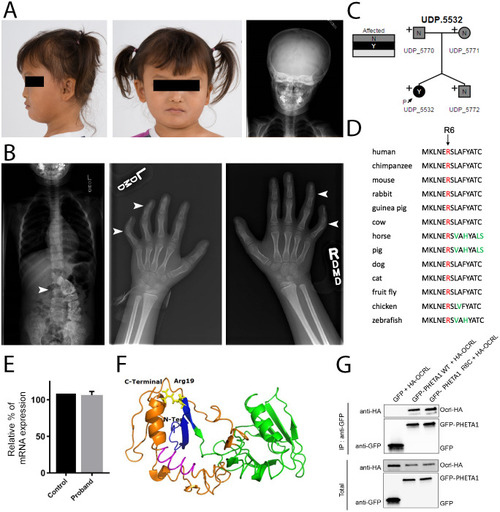
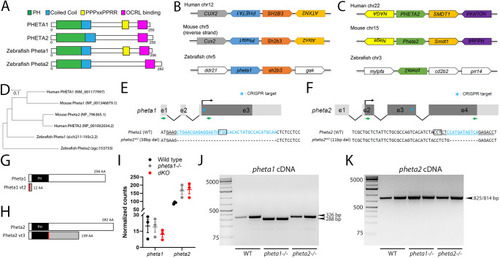
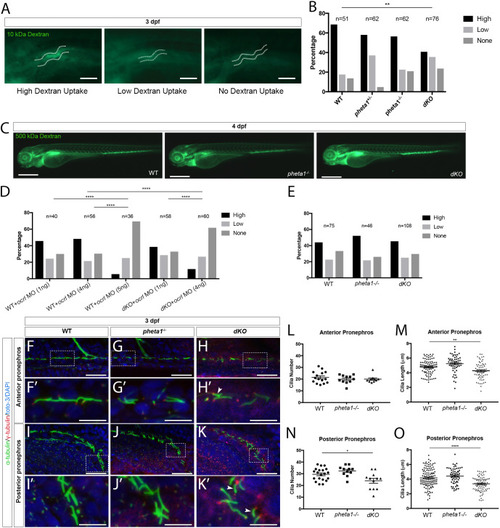
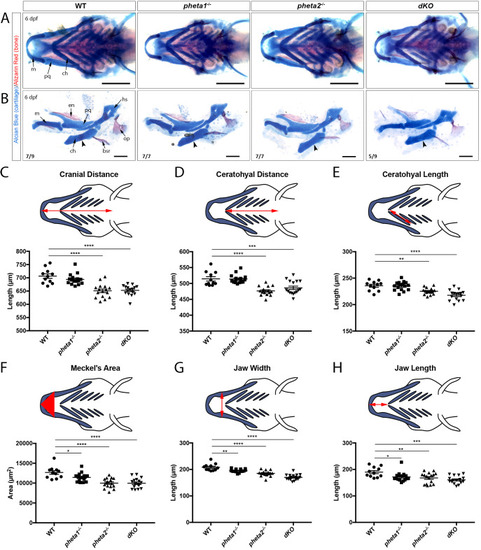
Deficiency in the endocytic adaptor proteins PHETA1/2 impair renal and craniofacial development
- Authors
- Ates, K.M., Wang, T., Moreland, T., Veeranan-Karmegam, R., Ma, M., Jeter, C., Anand, P., Wenzel, W., Kim, H.G., Wolfe, L.A., Stephen, J.A., Adams, D.R., Markello, T., Tifft, C.J., Settlage, R., Gahl, W.A., Gonsalvez, G.B., Malicdan, M.C., Flanagan-Steet, H., Pan, Y.A.
- Source
- Full text @ Dis. Model. Mech.
|
|
|
|
|
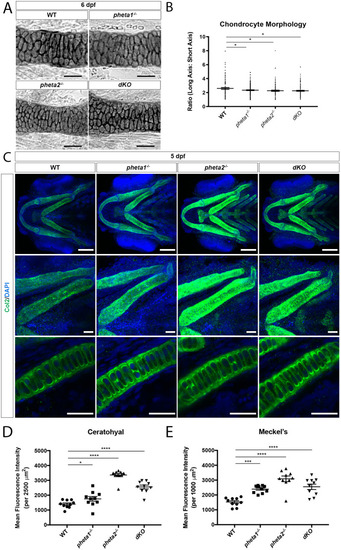
EXPRESSION / LABELING:
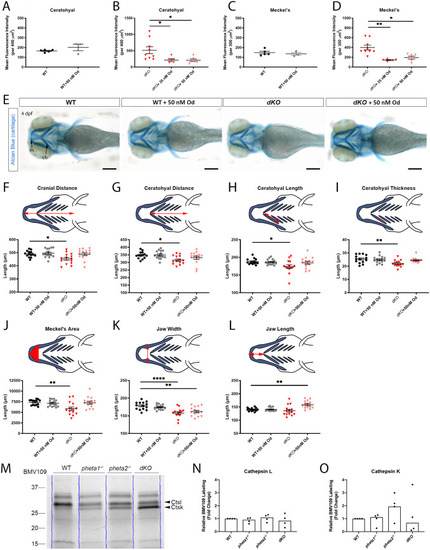
PHENOTYPE:
|
|
|
|
|
|
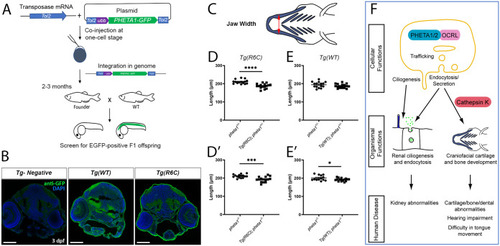
PHENOTYPE:
|
|
PHENOTYPE:
|