- Title
-
A genetic basis for molecular asymmetry at vertebrate electrical synapses
- Authors
- Miller, A.C., Whitebirch, A.C., Shah, A.N., Marsden, K.C., Granato, M., O'Brien, J., Moens, C.B.
- Source
- Full text @ Elife
|
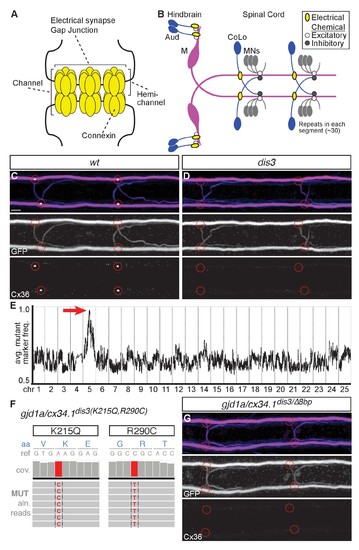
gjd1a/cx34.1 is required for electrical synapse formation. (A) Schematic of an electrical synapse, a specialized neuronal gap junction. (B) Simplified schematic of the zebrafish Mauthner (M) circuit in dorsal view with anterior to the left. Neurons and synapses of the hindbrain and two, of 30, spinal segments are shown. In the hindbrain mixed electrical/chemical synapses are made between auditory (Aud) afferent neurons and the M cell lateral dendrite (Aud/M electrical synapses). In the spinal cord the M axon makes electrical synapses with commissural local (CoLo) interneurons found in each segment (M/CoLo electrical synapses). (C,D) Two representative dorsal views of spinal cord segments from M/CoLo:GFP larvae at 5 days post fertilization. Images are maximum intensity projections of ~10 uM with anterior to the left. Scale bar = 10 uM. Larvae are stained for anti-GFP (magenta), anti-human-Connexin36 (Cx36, yellow), and for neurofilaments (RMO44, blue). Individual GFP and Cx36 channels are shown in neighboring panels. Associated experimental statistics are found in the figure-related table. The Cx36 staining found at wildtype M/CoLo synapses (C), red circles) is lost in dis3 mutants (D), red circles). (E) Genome wide RNA-seq-based mapping data. The average frequency of mutant markers (black marks) is plotted against genomic position. A single region on chromosome 5 (chr5) emerges with an allele frequency near 1.0 indicating linkage to the dis3 mutation (red arrow). Each chromosome is separated by vertical lines and labeled at the bottom. (F) Mutant reads from the RNA-seq mapping data at two separate positions within the gjd1a/cx34.1 gene are shown aligned to the reference genome identifying two missense changes within dis3 mutant animals. Wildtype reference (ref) genome nucleotides and encoded amino acids (aa) are noted. Aligned mutant (MUT) reads are shown as grey boxes; colored letters highlights differences from reference. (G) Electrical synapses are lost in trans-heterozygous animals carrying a dis3 and an 8 bp frameshift allele (dis3/8 bp) in gjd1a/cx34.1. Images of the spinal cord as in (C,D). Associated quantitation of Cx36 at wildtype or mutant synapses can be found in source data for Figure 1. |
|
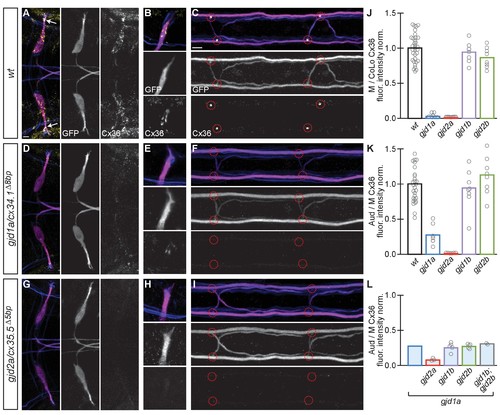
Electrical synapses of the Mauthner circuit require both gjd1a/cx34.1 and gjd2a/cx35.5. (A–I). In this and all subsequent figures, unless otherwise specified, images are dorsal views of hindbrain, Mauthner lateral dendrite, and two spinal cord segments from M/CoLo:GFP larvae at 5 days post fertilization. Hindbrain, lateral dendrite, and spinal cord images are maximum intensity projections of ~30,~5, and ~10 uM, respectively. Anterior is to the left. Scale bar = 10 uM. Larvae are stained for anti-GFP (magenta), anti-human-Connexin36 (Cx36, yellow), and for neurofilaments (RMO44, blue). Individual GFP and Cx36 channels are shown in neighboring panels. Electrical synapses are found on the Mauthner cell body (A) with prominent and stereotyped Aud/M synapses found on Mauthner’s lateral dendrite (arrows in A), (B) and at M/CoLo synapses in the spinal cord (C), red circles). (D–I) In gjd1a/cx34.1 and gjd2a/cx35.5 mutants Mauthner electrical synapses are lost or, in the specific case of Aud/M synapses in gjd1a/cx34.1 mutants, diminished. (J–L) Bar graphs represent the mean of the indicated value quantified at synapses with each circle representing the average of 12–16 M/CoLo or 8–12 Aud/M synapses within an animal. (J,K) M/CoLo electrical synapses are absent in gjd1a/cx34.1 and gjd2a/cx35.5 mutants, while Aud/M club endings are strongly reduced in gjd1a mutants and absent in gjd2a mutants. gjd1b and gjd2b have no effect on Mauthner electrical synapses. N = L. The remaining Cx36 staining observed at Aud/M synapses in gjd1a mutants is not due to gjd1b or gjd2b. For reference, the first bar is a duplication of gjd1a mutant data from K. In gjd1a;gjd2a double mutants the remaining Cx36 staining is lost as expected given that gjd2a is required for Aud/M synapses (K). In double and triple mutants combinations between gjd1a, gjd1b, and gjd2b there is no effect on the remaining Cx36 staining at Aud/M synapses. Associated quantitation of Cx36 at wildtype or mutant synapses can be found in Figure 3—source data 1 for Figure 3 and Figure 3—source data 2 for Figure 3. |
|
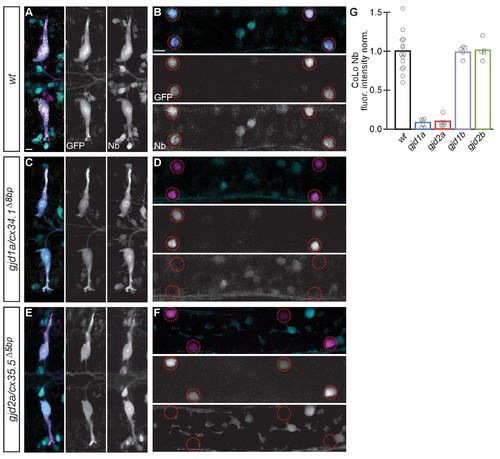
Electrical synapses are functionally defective in gjd1a/cx34.1 and gjd2a/cx35.5 mutants. Retrograde labeling of Mauthner axons with the gap junction permeable dye Neurobiotin (Nb) from a caudal transection. Hindbrain and spinal cord images are maximum intensity projections of ~30 and~10 uM, respectively. Anterior is to the left. Scale bar = 10 uM. Spinal cord images are at the level of the CoLo cell bodies (circles), which is dorsal to the synapses. (A–F) Larvae are stained for anti-GFP (magenta), biotin (Nb, cyan), and anti-human-Connexin36 (Cx36, yellow). Nb labels the Mauthner cell bodies and other caudally projecting neurons in the hindbrain (A) and passes from the Mauthner axon through the electrical synapses to fill the CoLo cell bodies (B), circles). Other neurons are also labeled due to projections caudally into the lesion site (A, non-Mauthner neurons, B, non-circled cell bodies). (C–F) In gjd1a/cx34.1 and gjd2a/cx35.5 mutants Nb labels M normally (C,E) however none passes into CoLos (D,F). (G) Quantitation of the ratio of Nb in CoLo to M cell bodies in wildtype and mutants. Each circle represents the average Nb fluorescence within 8–12 CoLo cell bodies compared to the 2 Mauthner cell bodies in an animal. Associated experimental statistics are found in the figure-related table. Associated quantitation of Nb transfer in wildtype or mutant can be found in source data for Figure 4. PHENOTYPE:
|
|
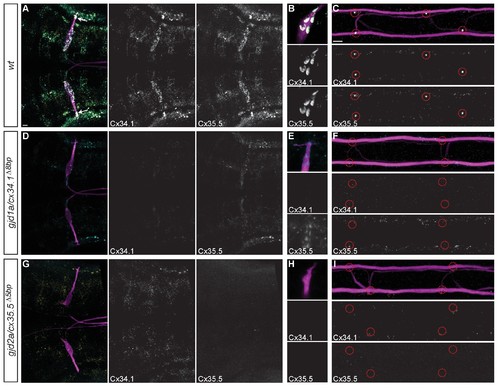
Recruitment of Gjd1a/Cx34.1 and Gjd2a/Cx35.5 to electrical synapses is co-dependent. Larvae are stained for anti-GFP (magenta), anti-Connexin34.1 (Cx34.1, yellow), and for anti-Connexin35.5 (Cx35.5, cyan). Individual Cx34.1 and Cx35.5 channels are shown in neighboring panels. Hindbrain, lateral dendrite, and spinal cord images are maximum intensity projections of ~30,~5, and ~10 uM, respectively. Anterior is to the left. Scale bar = 10 uM. (A–C) Cx34.1 and Cx35.5 are found colocalized at electrical synapses throughout the hindbrain (A) including at Aud/M (arrows in A, (B), and M/CoLo synapses (C). In gjd1a/cx34.1 mutants the localization of Cx35.5 to electrical synapses is lost (D–F) or diminished at Aud/M synapses (E); in gjd2a/cx35.5 mutants the localization of Cx34.1 to electrical synapses is lost (G–I). Associated antibody information can be found in source data for Figure 5. |
|
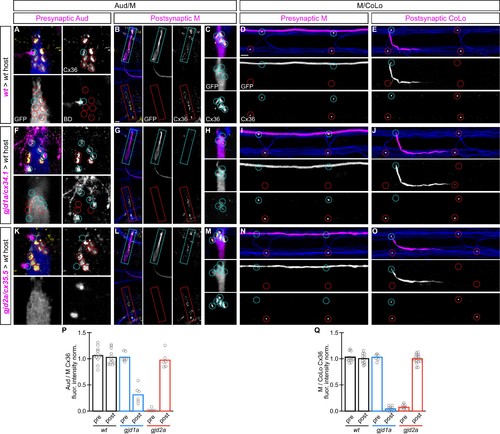
gjd1a/cx34.1 and gjd2a/cx35.5 are required asymmetrically at Mauthner electrical synapses. Dorsal views of chimeric larvae containing Biotin-Dextran- (BD) or GFP-marked cells transplanted from a donor embryo of noted genotype into a wildtype (wt) host; throughout the figure the neurons derived from the donor embryo are displayed in magenta, while those from the host are in blue. Synapses (stained with anti-human-Cx36, yellow) associated with a transplanted neuron (cyan circles and boxes) can be directly compared to wildtype host synapses (red circles and boxes). Hindbrain, lateral dendrite, and spinal cord images are maximum intensity projections of ~30,~5, and ~10 uM, respectively. Anterior is to the left. Scale bar = 10 uM. (A–C) At the Aud/M synapses presynaptic auditory afferent neurons (in A, stained with BD, magenta) synapse onto the postsynaptic Mauthner lateral dendrite (stained with anti-GFP, blue in A, magenta in B,C). (D,E) At the M/CoLo synapses the presynaptic Mauthner axon (stained with anti-GFP, magenta in D) synapses with the postsynaptic CoLo (stained with anti-GFP, magenta in E). (F–J) Presynaptic removal of gjd1a/cx34.1 function has no effect on Aud/M and M/CoLo synapses (cyan circles in F,I). By contrast, removing gjd1a/cx34.1 postsynaptically causes a loss of electrical synapse staining (cyan boxes and circles in G,H,J; note residual Cx36 staining at Aud/M synapses when gjd1a/cx34.1 is removed from only the postsynaptic neuron, (H). (K–O) Conversely, gjd2a/cx35.5 function is required exclusively presynaptically (K,N) and is dispensable postsynaptically (L,M,O). (P,Q) Quantitation of the ratio of Cx36 at donor-associated synapses to wildtype host synapses in chimaeric embryos. Each circle represents the average ratio of 1–8 donor-associated to 8–12 host-associated synapses within an animal, varying depending on the synapse and chimaera. Associated quantitation of Cx36 at chimeric synapses can be found in Figure 6—source data 1 for Figure 6 and Figure 6—source data 2 for Figure 6. |
|
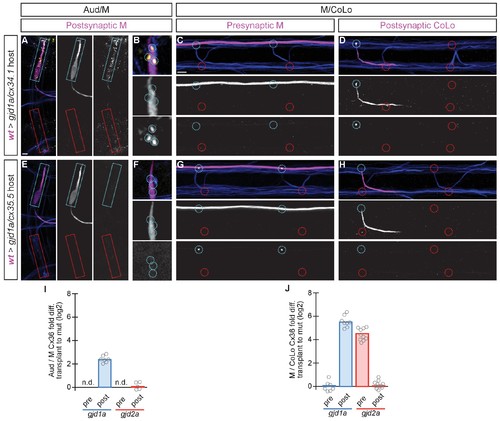
The exclusive asymmetric functions of gjd1a/cx34.1 and gjd2a/cx35.5 are sufficient for electrical synapse formation. Dorsal views of chimeric larvae containing GFP-marked cells transplanted from a wildtype (wt) donor embryo into a mutant host of noted genotype; throughout the figure the neurons derived from the wt donor embryo are displayed in magenta, while those from the mutant host are in blue. Synapses (stained with anti-human-Cx36, yellow) associated with a transplanted neuron (cyan circles and boxes) can be directly compared to mutant host synapses (red circles and boxes). Hindbrain, lateral dendrite, and spinal cord images are maximum intensity projections of ~30, ~5, and ~10 uM, respectively. Anterior is to the left. Scale bar = 10 uM. (A–D) When the postsynaptic neuron of the Aud/M or M/CoLo synapse is wildtype in a gjd1a/cx34.1 mutant the electrical synapses are rescued (A,B,D). By contrast, when the presynaptic neuron is wildtype in a mutant there is no effect on Cx36 staining at the synapse (C). (E–H) Conversely, electrical synapses are rescued in gjd2a/cx35.5 mutants only when the presynaptic, and not the postsynaptic, neuron is wildtype. (I,J) Quantitation of the fold increase in the ratio of Cx36 at wt-donor-associated synapses to mutant host synapses in chimaeric embryos. Each circle represents the average ratio of 1–8 donor-associated to 8–16 host-associated synapses within an animal. Associated quantitation of Cx36 at chimeric synapses can be found in Figure 7—source data 1 for Figure 7 and Figure 7—source data 2 for Figure 7. |
|
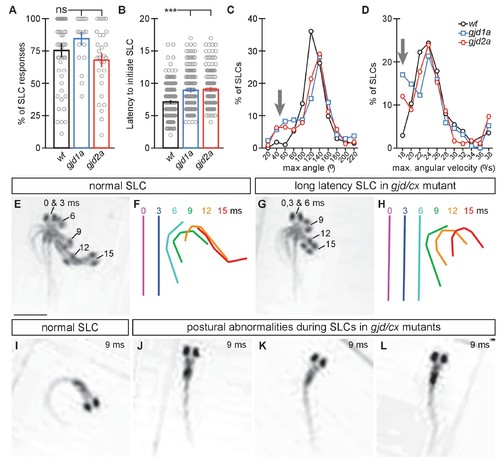
gjd1a/cx34.1 and gjd2a/cx35.5 mutants have delayed and abnormal escape responses. M-induced escape responses (Short Latency C-bends, SLCs) to a startling vibrational (sound) stimuli executed by 6 day post fertilization larvae were analyzed by high-speed (1000 frame per second) videomicroscopy. Scale bar = 1 mM. (A) Frequency of elicited SLCs in wildtype (wt) and indicated mutants in 10 trials per animal. Note that the FLOTE analysis software removes some trials if they cannot be classified. Bar graphs represent data as mean ± SEM with each circle representing an individual animal’s average % of response. Mutant larvae execute escapes as frequently as WT (n = 52, 20, and 29 larvae for wt, gjd1a/cx34.1, and gjd2a/cx35.5, respectively; 1 way ANOVA not significant (ns), Dunn’s Multiple Comparison Test: wt to gjd1a/cx34.1 and wt to gjd2a/cx35.5 both ns). (B) Latency of elicited SLCs in all individual trials. Bar graphs represent data as mean ± SEM with each circle representing individual SLC latencies. Mutant larvae are significantly delayed in their latency to initiate an M-induced SLC (n = 359, 160, and 180 SLCs from 52, 20, and 29 larvae from wt, gjd1a/cx34.1, and gjd2a/cx35.5, respectively; 1 way ANOVA p<0.0001, Dunn’s Multiple Comparison Test: wt to gjd1a/cx34.1 and wt to gjd2a/cx35.5 both significant at p<0.001). (C,D). Kinematic analysis of the maximum SLC turn angle (C) and angular velocity (D) plotted as the average number of events within an indicated bin. Arrows indicate shallow angle and low velocity turns exhibited by mutants. (E–H) Time-lapse analysis of a normal (E,F) and delayed (G,H) M-induced escape response (SLCs). Individual snapshots taken at the indicated times (ms = milliseconds) are overlaid on an individual image (E,G). A line representing the midline body axis at each time was drawn by hand to indicate the movement (F,H). (I–L) A normal escape bend at its maximum angle (I) compared to abnormally shaped escape bends executed by gjd1a (J,K) or gjd2a (L) mutant larvae. Associated experimental statistics can be found in source data for Figure 8. PHENOTYPE:
|