- Title
-
A simple strategy for heritable chromosomal deletions in zebrafish via the combinatorial action of targeting nucleases
- Authors
- Lim, S., Wang, Y., Yu, X., Huang, Y., Featherstone, M.S., and Sampath, K.
- Source
- Full text @ Genome Biol.
|
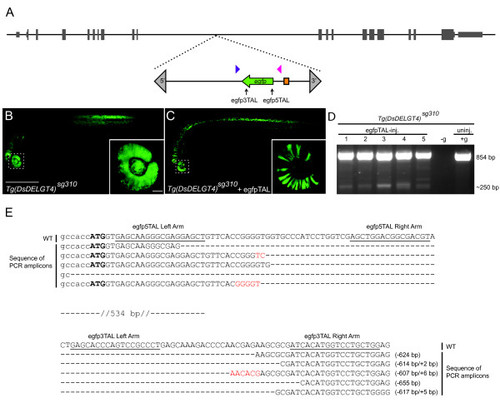
Targeted deletions in egfp. (a) Schematic representation of chromosome 21 showing the position of the Tg (Ds DELGT4) sg310 enhancer trap insertion. The Ds transposon terminal repeat sequences are indicated by grey triangles; green arrow indicates egfp reporter sequences and orientation, orange box indicates the glial fibrillary acidic protein mini-promoter; TALEN targeting sites are shown with black arrows and genotyping primers are indicated by blue and magenta triangles. (b) A 30 hours post-fertilization (hpf) Tg (Ds DELGT4) sg310 embryo showing ubiquitous and robust expression of EGFP; inset shows uniform EGFP expression in the eye; scale bars, 500 μm in (b); 50 μm in inset. (c) Tg (Ds DELGT4) sg310 embryo injected with egfp TALEN pairs showing patchy and reduced EGFP expression; inset shows loss of EGFP expression in some sectors of the eye. (d) PCR with primers spanning the TALEN targeting sites (blue and magenta triangles in (a)) shows the expected 250 bp truncated egfp and 854 bp full-length egfp products in individual embryos injected with egfp TALEN pairs, whereas only the full-length product is observed in the un-injected control embryo. No template control is indicated by -g. (e) Alignment of wild-type (WT) egfp sequences with mutated PCR amplicons shows various deletions of approximately 600 bp between the targeting sites, accompanied by small insertions (red). EXPRESSION / LABELING:
|
|
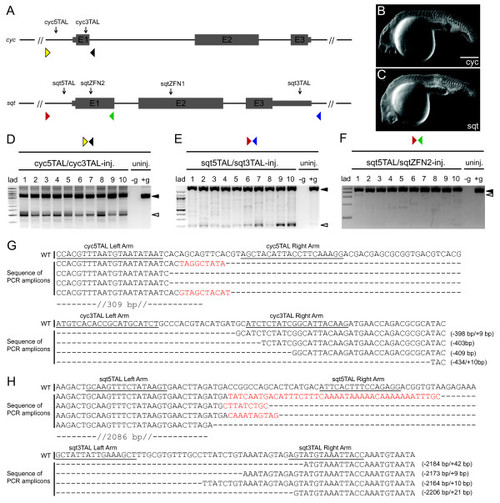
Targeted deletions in cyc and sqt by multiple TALEN and ZFN pairs. (a) Schematic representation of the cyc and sqt loci, with positions of the TALEN targeting sites and ZFN targeting sites indicated by black arrows. E1, E2 and E3 indicate cyc or sqt exons 1 to 3. Colored triangles in the both cyc and sqt panels indicate the position of primers used for genotyping. (b) Phenotype of cyc TALEN injected embryo at 24 hpf showing cyclopia. Scale bar, 100 µm. (c) Phenotype of representative sqt nuclease-injected embryo manifesting cyclopia and midline defects. (d) PCR with primers (yellow and black triangles in (a)) spanning the TALEN targeting sites (black arrows in (a)) shows the expected approximately 400 bp truncated cyc (white arrowhead), and 779 bp full-length cyc (black arrowhead) products in ten single embryos injected with cyc TALEN pairs, whereas the full-length product is observed in the un-injected control embryo. All embryos show faint intermediate sized products. No template control is indicated by -g. (e) PCR with primers (red and blue triangles in (a)) spanning the sqt locus show a 2.4 kb product (black arrowhead) for the intact sqt locus, whereas individual embryos with TALEN deletions show an approximately 220 bp complete locus deletion product (white arrowhead) and several other intermediate sized products. (f) PCR with primers spanning the sqt TSS site (red and green triangles in (a)) show a 478 bp full-length wild-type product (black arrowhead), and only one embryo (number 1) shows the expected approximately 300 bp deletion product (white arrowhead). (g) Alignment of wild-type (WT) cyc sequences with mutated PCR amplicons shows various deletions of approximately 400 bp between the targeting sites, accompanied by small insertions (red). (h) Alignment of wild type sqt sequences with mutated PCR amplicons shows various deletions of approximately 2.2 kb between the targeting sites, accompanied by small insertions (red). PHENOTYPE:
|
|
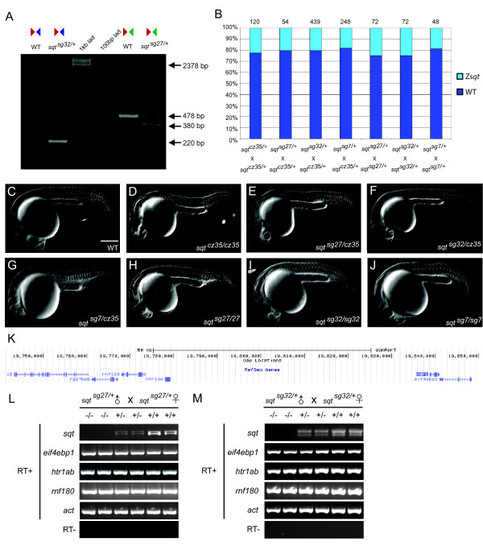
Heritable deletions in the sqt locus that result in RNA-null alleles. (a) PCR on single wild-type or sqt deletion mutant embryos (using primers indicated in Figure 2a) shows a 220 bp fragment in a sqtsg32 locus-deletion embryo, and a 380 bp fragment in TSS deleted sqtsg27 mutant embryo. Sometimes a larger approximately 500 bp fragment is observed in sqtsg27/+ heterozygous embryos, but the sequence is identical to the 478 bp product. (b) Percentage of embryos with sqt mutant phenotypes in sqt cz35/+, sqt sg27/+, sqt sg32/+ and sqt sg7/+ in-crosses and mating of sqt cz35/+ with sqt sg27/+, sqt sg32/+ and sqt sg7/+. The cz35 allele is an approximately 1.9 kb insertion in sqt exon 1; the sg27 allele is a 98 bp deletion of sqt TSS sequences; sg32 allele is a whole locus deletion of sqt; the sg7 ZFN allele harbors a GGCC insertion in sqt exon 2. (c-j) DIC images of 24 h wild-type (c), sqt cz35/cz35 (d), sqt sg27/cz35 (e), sqt sg32/cz35 (f), sqt sg7/cz35 (g), sqt sg27/sg27 (h), sqt sg32/sg32 (i), and sqt sg7/sg7 (j) embryos; scale bar in (c), 100 µm. (k) UCSC genome browser view of the sqt locus and neighboring genomic region. (l,m) RT-PCR with primers to detect expression of sqt RNA and transcripts of neighboring genes, eif4ebp1, rnf180, and htr1ab, shows lack of sqt RNA expression in sqt sg27/sg27 (l) and sqt sg32/sg32 (m) embryos whereas all neighboring gene transcripts are expressed at wild-type levels. Actin (act) expression was used as control. In contrast, both un-spliced and spliced sqt RNA is detected in wild-type and heterozygous embryos. EXPRESSION / LABELING:
PHENOTYPE:
|
|
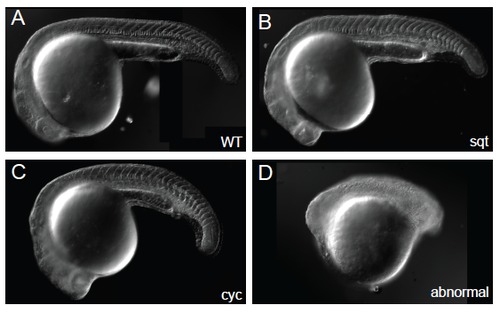
Representative phenotypes observed at 24 h in embryos injected with sqt or cyc TALENs. At high doses, the proportion of abnormal embryos increases. |