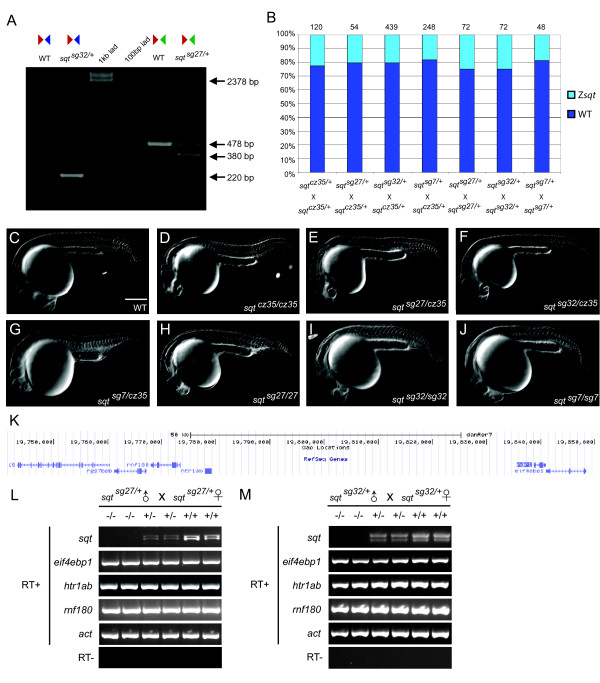
Fig. 3
Heritable deletions in the sqt locus that result in RNA-null alleles. (a) PCR on single wild-type or sqt deletion mutant embryos (using primers indicated in Figure 2a) shows a 220 bp fragment in a sqtsg32 locus-deletion embryo, and a 380 bp fragment in TSS deleted sqtsg27 mutant embryo. Sometimes a larger approximately 500 bp fragment is observed in sqtsg27/+ heterozygous embryos, but the sequence is identical to the 478 bp product. (b) Percentage of embryos with sqt mutant phenotypes in sqt cz35/+, sqt sg27/+, sqt sg32/+ and sqt sg7/+ in-crosses and mating of sqt cz35/+ with sqt sg27/+, sqt sg32/+ and sqt sg7/+. The cz35 allele is an approximately 1.9 kb insertion in sqt exon 1; the sg27 allele is a 98 bp deletion of sqt TSS sequences; sg32 allele is a whole locus deletion of sqt; the sg7 ZFN allele harbors a GGCC insertion in sqt exon 2. (c-j) DIC images of 24 h wild-type (c), sqt cz35/cz35 (d), sqt sg27/cz35 (e), sqt sg32/cz35 (f), sqt sg7/cz35 (g), sqt sg27/sg27 (h), sqt sg32/sg32 (i), and sqt sg7/sg7 (j) embryos; scale bar in (c), 100 µm. (k) UCSC genome browser view of the sqt locus and neighboring genomic region. (l,m) RT-PCR with primers to detect expression of sqt RNA and transcripts of neighboring genes, eif4ebp1, rnf180, and htr1ab, shows lack of sqt RNA expression in sqt sg27/sg27 (l) and sqt sg32/sg32 (m) embryos whereas all neighboring gene transcripts are expressed at wild-type levels. Actin (act) expression was used as control. In contrast, both un-spliced and spliced sqt RNA is detected in wild-type and heterozygous embryos.