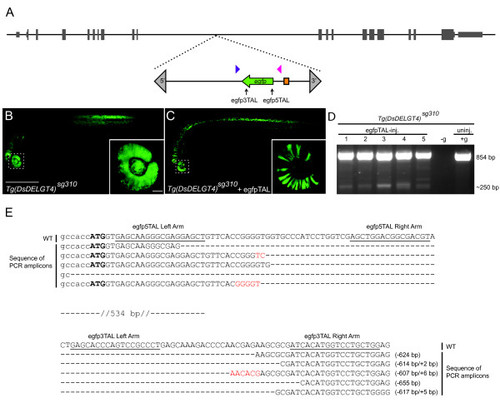
Targeted deletions in egfp. (a) Schematic representation of chromosome 21 showing the position of the Tg (Ds DELGT4) sg310 enhancer trap insertion. The Ds transposon terminal repeat sequences are indicated by grey triangles; green arrow indicates egfp reporter sequences and orientation, orange box indicates the glial fibrillary acidic protein mini-promoter; TALEN targeting sites are shown with black arrows and genotyping primers are indicated by blue and magenta triangles. (b) A 30 hours post-fertilization (hpf) Tg (Ds DELGT4) sg310 embryo showing ubiquitous and robust expression of EGFP; inset shows uniform EGFP expression in the eye; scale bars, 500 μm in (b); 50 μm in inset. (c) Tg (Ds DELGT4) sg310 embryo injected with egfp TALEN pairs showing patchy and reduced EGFP expression; inset shows loss of EGFP expression in some sectors of the eye. (d) PCR with primers spanning the TALEN targeting sites (blue and magenta triangles in (a)) shows the expected 250 bp truncated egfp and 854 bp full-length egfp products in individual embryos injected with egfp TALEN pairs, whereas only the full-length product is observed in the un-injected control embryo. No template control is indicated by -g. (e) Alignment of wild-type (WT) egfp sequences with mutated PCR amplicons shows various deletions of approximately 600 bp between the targeting sites, accompanied by small insertions (red).
|

