- Title
-
Deep Brain Photoreceptors Control Light-Seeking Behavior in Zebrafish Larvae
- Authors
- Fernandes, A.M., Fero, K., Arrenberg, A.B., Bergeron, S.A., Driever, W., and Burgess, H.A.
- Source
- Full text @ Curr. Biol.
|
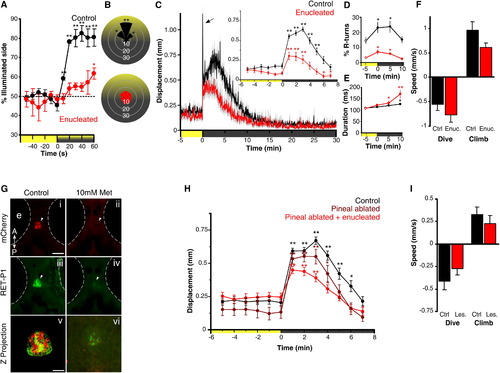
Light-Driven Behavior in Larval Zebrafish without Eyes or Pineal(A) Attraction of control and enucleated larvae to a phototaxis stimulus, measured by the percent of larvae observed on the illuminated side of the testing arena over time. Enucleated larvae exhibit a gradual shift to the illuminated side of the arena (symbols show one-sample t test to 50%; n = 4 groups of 15 larvae). Larval positions were recorded every second and then averaged over 10 s for each time point. Color along x axis indicates light condition.(B) Larval body orientation during exposure to a phototaxis stimulus. A significant proportion of control larvae exhibit a “head-on” orientation toward the spotlight (one-way ANOVA; F7, 24 = 51.21, p < 0.001; comparisons are Tukey’s post hoc), whereas enucleated larvae show no bias in body orientation (ANOVA; F7, 24 = 1.73, p = 0.15; n = 4 groups of 15 larvae). Data represent mean proportion of larvae oriented relative to the target light over 1 min.(C) Locomotor activity during dark-induced VMR. Arrow indicates O-bend spike observed only in controls. Inset: enucleated larvae significantly increase activity following light extinction (repeated-measures ANOVA; F2.5, 88.7 = 16.57, p < 0.001; n = 36 larvae). Data represent the mean activity for the preceding minute. Color along x axis indicates light condition. Pairwise comparisons are to the baseline time point at 5 min.(D and E) Kinematic analysis of VMR. Enucleated larvae (red) retain elevated R-turn initiation frequency (D; repeated-measures ANOVA; F2.1, 56.6 = 4.63, p = 0.013) and swim bout duration (E; repeated-measures ANOVA; F3, 40 = 6.41, p = 0.001) as seen in controls. Data represent the mean of observations during the first 16 s following each time point. Pairwise comparisons are to the baseline measurement at 5 min (empty circles) (control: n = 18 groups of 10 larvae; enucleated: n = 28 groups of 10 larvae).(F) Diving and climbing speed during VMR. In either response, enucleated larvae were not significantly different from controls (dive: t test, p = 0.27; climb: t test, p = 0.07; n = 6 groups of 5 larvae). Additionally, speed in all conditions is significantly different from 0 (one-sample t test, p < 0.005). Data represent mean vertical swim speed over the first 20 s of dive and ascent.(G) Nitroreductase-mediated ablation of the pineal. Dorsal views at 6 days postfertilization (dpf) are shown. (i–iv) Epifluorescence images of the pineal (arrow) in untreated and metronidazole (Met)-treated Tg(tph2:NfsB-mCherry)y227 larvae with anti-mCherry (red; i and ii) and anti-RET-P1 (green; iii and iv). Scale bar represents 100 μm. (v and vi) Confocal z projections (mCherry + RET-P1) showing concurrent nitroreductase and opsin expression in the pineal in untreated (v) and Met-treated (vi) larvae. Scale bar represents 25 μm.(H) VMR in enucleated, pineal-ablated larvae. Both pineal-ablated and pineal-ablated + enucleated larvae show a robust VMR following light extinction (repeated-measures ANOVA; F3.8,131.3 = 32.44, p < 0.001) (control and pineal-ablated + enucleated: n = 36 larvae; pineal-ablated: n = 26 larvae). Data represent mean activity for the preceding minute. Pairwise comparisons are to baseline time point 5 min.(I) Diving and climbing speed of enucleated, pineal-ablated larvae during VMR. In either response, lesioned larvae were not significantly different from controls (t test; dive: p = 0.26; climb: p = 0.42). Mean speed of dive and ascent for both groups is significantly different from zero (one-sample t test, p < 0.005; n = 14 groups of 5 larvae). Data represent mean swim speed over the first 20 s of dive and ascent.For all panels, error bars show SEM; p < 0.05, p < 0.01. See also Figure S1. EXPRESSION / LABELING:
|
|
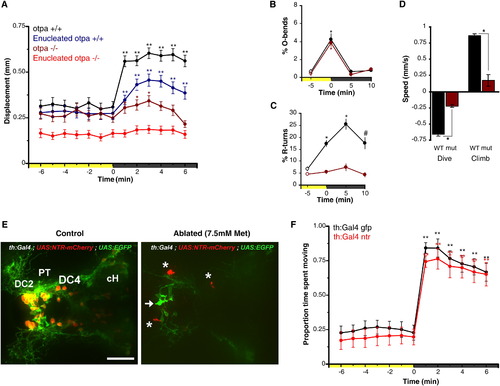
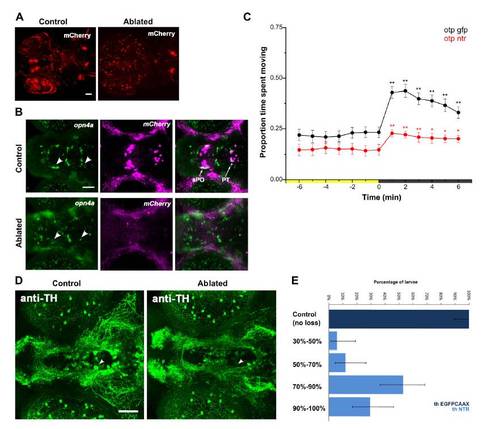
Reduction of VMR in otpa Mutants and Lack of Dopaminergic Contribution(A) Locomotor activity during dark-induced VMR of intact and enucleated otpa mutants and sibling larvae. Intact mutants show a response to light extinction (repeated-measures ANOVA; F3.0,175 = 8.5, p < 0.01; n = 59 larvae) that is greatly reduced relative to controls (intact siblings: n = 47 larvae; enucleated siblings: n = 97 larvae). Without eyes, mutants lose any response to light extinction (repeated-measures ANOVA; F3.2,317 = 1.72, p = 0.16; n = 101 larvae). Data represent mean activity for the preceding minute. Color along x axis indicates light condition. Pairwise comparisons to baseline time point at 0 min: p < 0.05, p < 0.01.(B and C) Kinematic analysis of photokinesis in intact otpa mutants. Otpa mutants retain O-bend responses to light extinction (B) (repeated-measures ANOVA; F3,9 = 53.7; p < 0.001; n = 4 groups of 10 larvae) but do not show characteristic increases in R-turn initiation (C) (mutants, red; siblings, black) (F3,9 = 2.1; p = 0.17; n = 4 groups of 10 larvae). #p < 0.05, p < 0.01 for pairwise comparisons to baseline at 5 min (empty circles). Data represent the mean and SEM of observations during the first 16 s following each time point.(D) Diving and climbing speed of intact otpa mutant larvae. Compared to siblings, otpa mutants exhibit significantly reduced diving speed during and climbing speed following a 60 s dark flash (t test; dive: p < 0.001; climb: p < 0.005; n = 3 groups of 8 larvae). Data represent mean and SEM swim speed over first 20 s of dive and ascent.(E) Nitroreductase-mediated ablation of dopaminergic (DA) neurons in Tg(BACth:Gal4VP16)m1233; Tg(UAS:EGFPCAAX); Tg(UAS-E1b:NfsB-mCherry) triple-transgenic larvae. Asterisk indicates mCherry aggregates remaining from ablated cells. Arrow indicates GFP-expressing nonablated cells. The following abbreviations are used: DC2 and DC4, Otp-dependent dopaminergic groups 2 and 4; PT, posterior tuberculum; cH, caudal hypothalamus. Dorsal view is shown. Scale bar represents 50 μm.(F) VMR in DA neuron-ablated larvae. Control Tg(th:Gal4VP16); Tg(UAS:EGFPCAAX) (black line) and DA neuron-ablated Tg(th:Gal4VP16); Tg(UAS-E1b:NfsB-mCherry) (red line) larvae show similar, robust VMR following light extinction (repeated-measures ANOVA; F12, 564 = 148.29, p < 0.001; n = 48 larvae). Data represent mean and SEM activity as in (A). Pairwise comparisons to baseline time point at 5 min: p < 0.05, p < 0.01.See also Figure S2. |
|
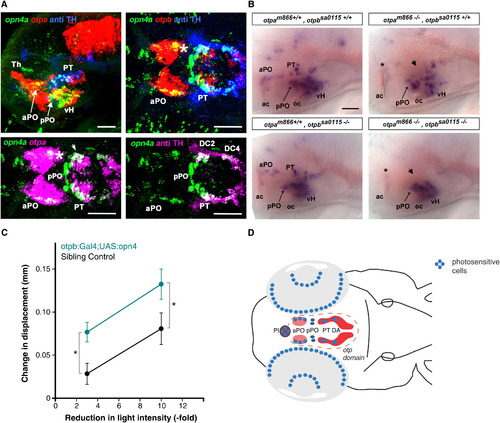
opn4a Expression Depends on Otp Activity in Areas of Coexpression with otpa(A) Analysis of expression domains of opn4a, otpa, and TH (anti-TH) in 3 dpf wild-type larvae. Top row shows z projections of combined channels (see labels) of confocal stacks recorded from lateral (left) and dorsal (right) views of the brain. Anterior is at left; dorsal is at top for lateral view. Bottom row shows z projections of channel combinations of a confocal stack showing dorsal views of the brain. Scale bars represent 50 μm. opn4a is coexpressed with otpa in the anterior preoptic area (aPO) (asterisk) and posterior tuberculum (PT) (arrowhead). The following abbreviations are also used in (A), (B), and (D): Th, thalamus; pPO, posterior preoptic area; vH, ventral hypothalamus; DC2, dopaminergic group 2; DC4, dopaminergic group 4; ac, anterior commissure; oc, optic chiasm; PI, pineal; PT DA, posterior tuberculum dopaminergic neurons.(B) Expression of opn4a in otpa and otpb mutants. Whole-mount in situ hybridization reveals loss of opn4a expression in the aPO (asterisk) and PT (arrowhead) of otpa and otpa, otpb double mutants (3 dpf). otpb mutants alone did not significantly affect opn4a expression, which is in line with the previously reported compensation of otpb knockdown by otpa activity in A11 DA neuron differentiation [20]. Anterior is at left; dorsal is up. Scale bar represents 50 μm.(C) Increase in activity following decrements in light intensity in enucleated Tg(otpb.A:Gal4)zc67; Tg(UAS:GFP-v2a-opn4)y233 larvae. Data show the difference in mean activity between 2 min after light change and 1 min prior to light change (mean displacement at t2 mean displacement at t1). Enucleated opn4-overexpressing larvae show an increased response to decrements in light intensity (repeated-measures two-way ANOVA; F1,100 = 8.84, p < 0.005; nonexpressing control: 28 larvae; GFP positive in otpb domain: n = 42 larvae). p < 0.001.(D) Schematic summarizing our results suggesting that preoptic opn4a-expressing neurons are deep brain photoreceptors driving dark photokinesis. We eliminated all depicted photoreceptive regions except the PO and found that the VMR response remained intact. Because otpa mutants lack aPO but not pPO opn4a expression, and because mutants without eyes do not show a VMR, the behavior must originate in the opn4a-positive cells in the aPO (pink). The illustrated DA domain (red) only comprises the diencephalic groups 2–6.See also Figure S3. EXPRESSION / LABELING:
|
|
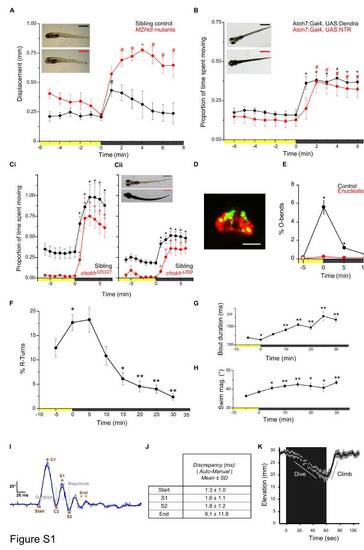
Characterization of VMR, Related to Figure 1 |
|
VMR in Larvae with Ablated Ventral Diencephalic Otp-Expressing Neurons and DA Neuron Ablation Efficiency, Related to Figure 2 |
|
Expression of Nonvisual Opsins in the otpa Domain and Expression of opn4a in Wild-Type and otp Mutant Embryos, Related to Figure 3 EXPRESSION / LABELING:
|