- Title
-
Melanophore Migration and Survival during Zebrafish Adult Pigment Stripe Development Require the Immunoglobulin Superfamily Adhesion Molecule Igsf11
- Authors
- Eom, D.S., Inoue, S., Patterson, L.B., Gordon, T.N., Slingwine, R., Kondo, S., Watanabe, M., and Parichy, D.M.
- Source
- Full text @ PLoS Genet.
|
Defective adult pigment stripes in seurat mutants. Shown are adult fish (A, B, C) and details of patterns (A2, B2, C2). (A, A2) Wild-type fish exhibit several dark stripes of melanophores and iridophores, as well as light “interstripes” of xanthophores and iridophores. (B, B2) Homozygous seuratutr15e1 mutants exhibit irregular spots of melanophores. (C, C2) seuratwp15e2/seuratutr15e1 fish exhibit a less severe stripe defect, most evident ventrally (arrowhead). seuratwp15e3/seuratutr15e1 exhibit a phenotype indistinguishable from seuratwp15e2/seuratutr15e1. At this stage, seuratwp15e2 and seuratwp15e3 are nearly indistinguishable from the wild-type when homozygous, though phenotypes are more apparent during the initial stages of stripe formation (not shown). Scale bar: in (A2), 1 mm for (A2–C2). PHENOTYPE:
|
|
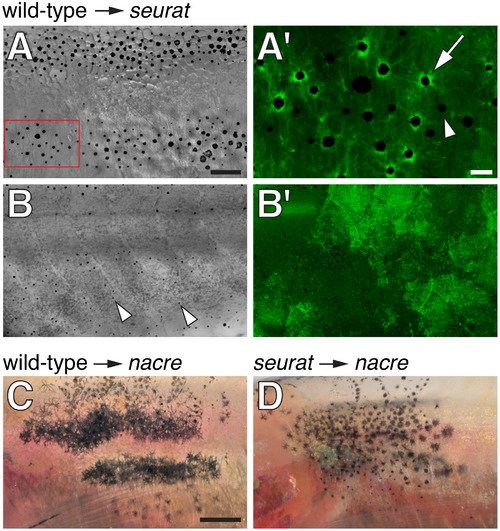
seurat is required autonomously to the melanophore lineage. (A, B) Wild-type Tg(bactin:GFP) cells transplanted to seurat mutant hosts. Fish shown are juveniles (~13 mm standardized standard length, SSL [11]) and were treated just prior to imaging with epinephrine, which contracts melanosomes towards cell bodies, thereby facilitating the detection of GFP fluorescence. (A) Chimeras that developed wild-type melanophores exhibited patches of restored stripes (n = 6). (A2) Detail of boxed region in A, showing GFP+ melanophores (e.g., arrow), as well as occasional GFP, presumptive seurat mutant melanophores (e.g., arrowhead). (B) Chimeras in which wild-type melanophores failed to develop exhibited a seurat mutant pattern of dispersed melanophores (arrowheads; n>100). In the example shown here, wild-type GFP+ cells developed as epidermis (B2 shown at same magnification as B). (C) When wild-type melanophores differentiated in a nacre mutant background, patches of normal stripes developed (n = 3; [16]). (D) By contrast, when seurat mutant melanophores differentiated in nacre hosts, these cells retained a dispersed pattern, as in the seurat mutant (n = 8), indicating a failure of xanthophores, iridophores, or other cell types to rescue melanophore stripe organization. In additional experiments, in which nacre; Tg(bactin:GFP) cells were transplanted to seurat mutant hosts, the differentiation of nacre- GFP+ (seurat+) iridophores likewise failed to rescue melanophore stripes in the seurat mutant background (donor xanthophores did not develop in these chimeras; data not shown). Scale bars: in (A) 100 μm for (A,B,B2); in (A2) 20 μm for (A2); in (C) 500 μm for (C,D). |
|
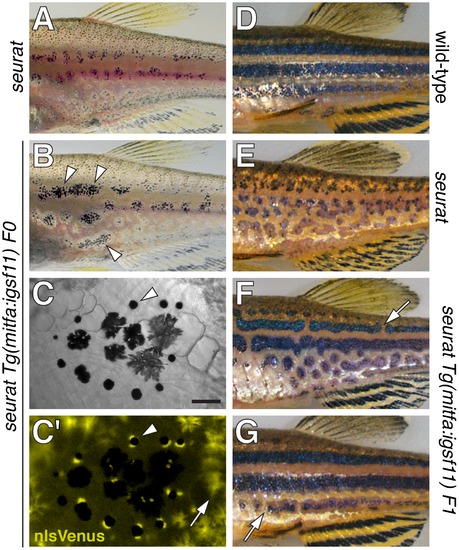
seurat mutant phenotype rescued by wild-type igsf11 expressed in mitfa+ pigment cell precursors. (A) Juvenile seurat mutant. (B) Sibling of fish in A injected with mitfa:nlsVenus-V2a-igsf11 transgene at the one cell stage exhibits patches of rescued melanophore stripes (arrowheads). (C,C2) Close-up showing individual melanophores (e.g., arrowhead) that express nlsVenus. This individual had been treated with epinephrine to contract melanin-containing organelles (melanosomes) towards the cell body, which facilitates fluorophore visualization; melanosomes of only some melanophores are in the fully contracted state. Arrow, xanthophores autofluorescence in the same channel as nlsVenus. (D) Adult wild-type. (E) Adult seurat mutant. (F, G) Two examples of seurat mutants transgenic for stably integrated mitfa:igsf11, which exhibit nearly wild-type stripes marked by occasional breaks (arrows). Individuals were genotyped by PCR to verify presence of the transgene. Scale bar: in (C) 60 μm for (C,C2). PHENOTYPE:
|
|
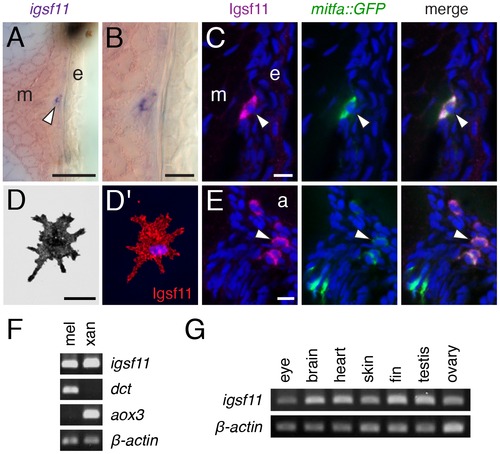
igsf11 is expressed by pigment cells and their precursors. (A,B) In situ hybridization for igsf11 transcript during the larval-to-adult transformation, showing an igfs11-expressing cell near the hypodermis (A, and higher magnification in B). e, epidermis; m, myotome. (C) Similar location to that shown in (B), illustrating a cell within the hypodermis (arrowhead), coexpressing Igsf11 cell (magenta) and mitfa:GFP (green). Nuclei in all immunofluorescence images are counterstained with DAPI (blue). (D,D2) A melanophore isolated in vitro expresses Igsf11 (red). (E). Extra-hypodermal Igsf11+ cells (arrowhead) also coexpressed Igsf11 (magenta) and mitfa:GFP, though some mitfa:GFP+ cells were Igsf11 (lower left of panels). Shown here are cells just ventral to the aorta (a). (F) RT-PCR showed that cell populations isolated by differential centrifugation and highly enriched for melanophores (mel) and xanthophores (xan) express igsf11 transcript. dct, dopachrome tautomerase, expressed by melanophores; aox3, aldehyde oxidase 3, expressed by xanthophores. β-actin, loading control. (G) igsf11 expression was detected in several additional tissue types dissected from adult fish. Scale bars: in (A) 40 μm for (A); in (B) 10 μm for (B); in (C) 10 μm for (C,E); in (D) 20 μm for (D). EXPRESSION / LABELING:
|
|
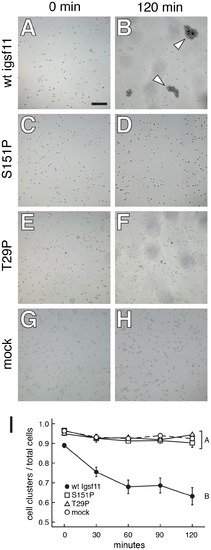
Igsf11 promotes aggregation of K562 myeloid leukemia cells in vitro. Cells were transfected with wild-type zebrafish igsf11 (A, B), S151P, seuratutr15e1 mutant igsf11 (C, D), T29P, seuratwp15e2 mutant igsf11 (E, F), or mock transfected (G, H). At the start of the experiment, the numbers of small cellular aggregates and total numbers of cells were similar (A, C, E, G). By 120 min, a relatively small number of aggregates containing numerous cells had formed in the cells transfected with wild-type igsf11 (arrowheads in B), though this was not the case in cells of other treatments (D, F, H). (I) Quantitation of the ratio of cellular aggregates to the total number of cells (mean±SE) confirmed that cells were increasingly found in fewer, larger aggregates when transfected with wild-type igsf11. At 120 min, degrees of aggregation differed significantly among treatments overall (F3,48 = 19.8, P<0.0001). Post hoc means comparisons indicated that aggregation behavior in cells transfected with wild-type igsf11 (B in the figure) differed significantly from that of cells transfected with mutant igsf11 or controls (A in the figure; Tukey Kramer post hoc comparisons, P<0.01); aggregation of cells transfected with mutant forms of igsf11 did not differ significantly from one another or from mock transfected cells. Values shown are least squares means adjusted to control for variation after controlling for minor but significant variation among replicates (P<0.01). |
|
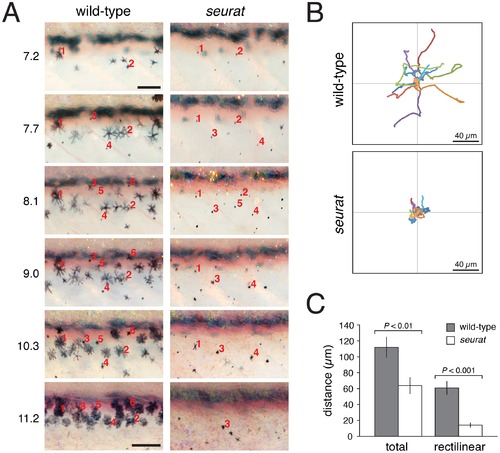
igsf11-dependent migration and survival of melanophores. (A) Repeated images of developing wild-type and seurat mutant larvae between 14–28 days post-fertilization. Numbers to the left of images are SSL. In wild-type larvae, new adult melanophores differentiated already within stripes or translocated short distances as stripes formed (e.g., note changes in the relative positions of cells 2 vs. 4, and cell 3 vs. 1 and 5). In seurat mutants, however, little movement was observed and many melanophores died as evidenced by the presence of melanized cellular debris apparent at high magnification (not shown; [32]. Images shown were rescaled to maintain the same field of view as the fish grew; scale bars at 7.2 SSL and 11.2 SSL represent 100 and 200 μm, respectively. (B) When cultured in vitro, wild-type melanophores migrated further than seurat mutant melanophores. Shown are tracks of 16 cells of each genotype. (C) Quantification of total and rectilinear distances moved by cells in vitro confirmed reduced motility of seurat mutant melanophores (t = 3.0, t = 5.4, respectively; d.f. = 26). Shown are means ± SE. PHENOTYPE:
|
|
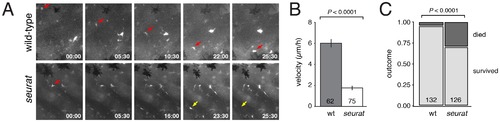
Melanophore precursors require igsf11 for their migration and survival. (A) Selected frames from time-lapse movies of mitfa:GFP+ cells in wild-type and seurat mutant explants. A single cell (red arrow) moved from dorsal to ventral over the duration of the movie. In a seurat mutant, many cells failed to migrate (e.g., red arrow) or died (yellow arrow) during the period of imaging. (B) Velocities (mean±SE) of mitfa:GFP+ cells were significantly reduced in seurat mutants compared to the wild-type (t = 11.2, d.f. = 135), as were total distances traveled (not shown). (C) seurat mutant melanophores were also significantly more likely to die than were wild-type melanophores (X2 = 29.8, d.f. = 1). |
|
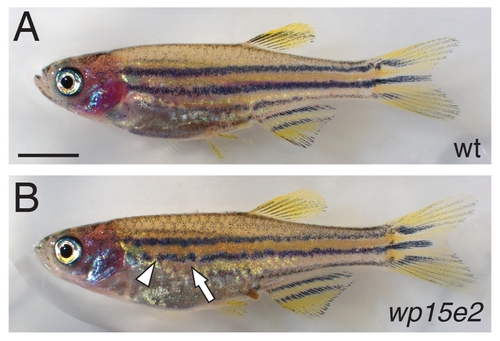
Developmental phenotype of weak allele, seuratwp15e2. (A) Wild-type juvenile. (B) Homozygous seuratwp15e2 juvenile. Arrow, typical irregularity in melanophore stripe border. Arrowhead, break in melanophore stripe. Scale bar: in (A) 5 mm for (A,B). PHENOTYPE:
|
|
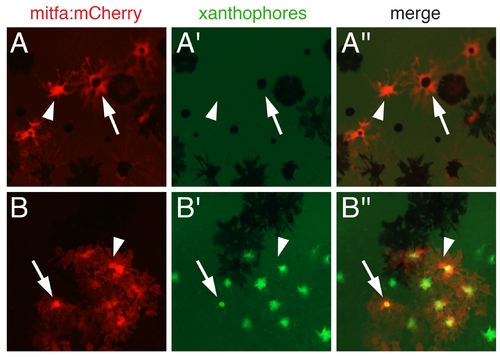
mCherry expression driven by the mitfa promoter. Shown is expression from a 1.3 kb fragment of the mitfa promoter in mosaic, transiently transgenic late larval fish (~9.5 SSL). (A) mitfa:mCherry was expressed by newly differentiated melanophores (arrow) as well as undifferentiated cells that may be precursors to melanophores and xanthophores (arrowhead). (B) Expression of mitfa:mCherry in differentiated xanthophores, which autofluoresce in the GFP channel (arrow), as well as in undifferentiated cells (arrowhead). Expression from a 2.2 kb fragment of the mitfa promoter was similar to that shown here. |
|
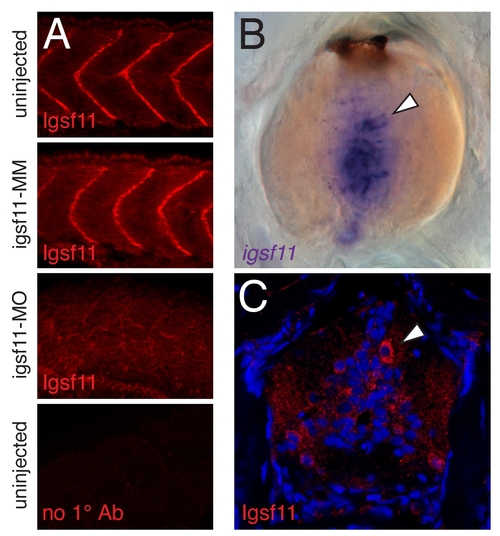
Characterization of Igsf11 antiserum. (A) Knockdown by morpholino oligonucleotide injection reduced Igsf11 immunoreactivity in embryos at 24 hours post-fertilization. Igsf11 immunoreactivity was present along vertical myosepta in uninjected embryos as well as embryos injected with a control igsf11 5 bp mismatch morpholino (igsf11-MM), but was dramatically reduced in embryos injected with a morpholino targeting the igsf11 translational start site (igsf11-MO). Embryos were injected with 4 ng of either morpholino and exposure times were identical for all images shown. (B,C) In addition to scattered cells in the hypodermis and extra-hypodermal locations (main text), both in situ hybridization (B) and immunohistochemistry (C) revealed igsf11-expressing cells (arrowheads) in the spinal cord during the larval-to-adult transformation (larvae shown here at ~9 SSL [11]). Staining appears more extensive in B than C owing to different section thicknesses (150 µm, 20 µm, respectively). EXPRESSION / LABELING:
|
|
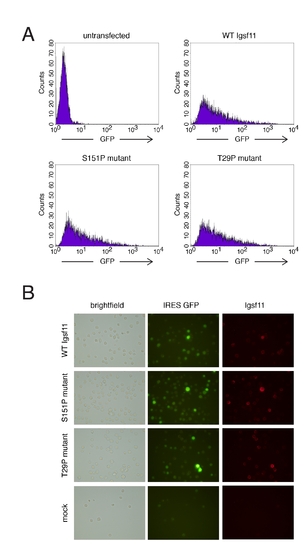
Transfection efficiency and expression of wild-type and mutant Igsf11 by K562 human myeloid leukemia cells. (A) Fluorescence activated cell sorting indicated similar transfection efficiencies for cells transfected with wild-type or mutant forms of Igsf11. (B) Immuncytochemistry confirmed expression of wild-type and mutant forms of Igsf11 by K562 cells (shown here without rotary culturing or aggregration). Mock treated cells were transfected with pIRES2-AcGFP1vector alone. |