- Title
-
Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling
- Authors
- Chaki, M., Airik, R., Ghosh, A.K., Giles, R.H., Chen, R., Slaats, G.G., Wang, H., Hurd, T.W., Zhou, W., Cluckey, A., Gee, H.Y., Ramaswami, G., Hong, C.J., Hamilton, B.A., Cervenka, I., Ganji, R.S., Bryja, V., Arts, H.H., van Reeuwijk, J., Oud, M.M., Letteboer, S.J., Roepman, R., Husson, H., Ibraghimov-Beskrovnaya, O., Yasunaga, T., Walz, G., Eley, L., Sayer, J.A., Schermer, B., Liebau, M.C., Benzing, T., Le Corre, S., Drummond, I., Janssen, S., Allen, S.J., Natarajan, S., O'Toole, J.F., Attanasio, M., Saunier, S., Antignac, C., Koenekoop, R.K., Ren, H., Lopez, I., Nayir, A., Stoetzel, C., Dollfus, H., Massoudi, R., Gleeson, J.G., Andreoli, S.P., Doherty, D.G., Lindstrad, A., Golzio, C., Katsanis, N., Pape, L., Abboud, E.B., Al-Rajhi, A.A., Lewis, R.A., Omran, H., Lee, E.Y., Wang, S., Sekiguchi, J.M., Saunders, R., Johnson, C.A., Garner, E., Vanselow, K., Andersen, J.S., Shlomai, J., Nurnberg, G., Nurnberg, P., Levy, S., Smogorzewska, A., Otto, E.A., and Hildebrandt, F.
- Source
- Full text @ Cell
|
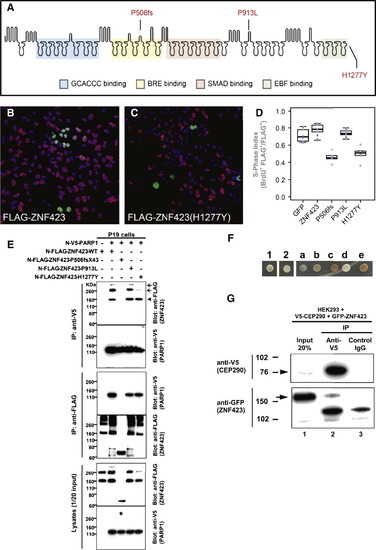
Two ZNF423 Mutations Have Dominant Negative Characteristics, ZNF423 Mutation Abrogates Interaction with PARP1, and ZNF423 Directly Interacts with the NPHP-RC Protein CEP290/NPHP6 (A) Amino acid residues altered by NPHP-RC mutations in ZNF423 are drawn in relation to functional annotation of its 30 Zn-fingers. (B–D) S-phase index assay (fraction of transfected cells incorporating BrdU) for P19 cells transfected with either wild-type or mutant ZNF423. (B) Representative field of cells transfected with wild-type ZNF423 shows high frequency of BrdU+ FLAG+ double-positive cells. (C) ZNF423–H1277Y transfected cells exhibits fewer FLAG–positive cells and a lower proportion that are double positive. (D) S-phase index measured in duplicate transfections for each of three DNA preparations per construct. A GFP construct was used as a nonspecific control. Constructs with P506fsX43 and H1277Y mutations detected in NPHP-RC show significantly reduced S-phase index (p < 105, ANOVA with post-hoc Tukey honestly significant difference [HSD]). (E) ZNF423 interacts with PARP1. P19 cells were cotransfected with expression constructs for N terminally FLAG-tagged human full-length ZNF423 and V5-tagged human full length PARP1. Comparable amounts of both proteins were present in all lysates (lower panels). Proteins were precipitated, using anti-V5 (upper panels) and anti-FLAG antibodies (middle panels), respectively. Reciprocal coIP demonstrates interaction between ZNF423 and PARP1. Note that the ZFN423 mutation P506fsX43 abrogates this interaction (arrowhead) and that mutation H1277Y diminishes ZNF423 multimerization (arrow). (F–G) ZNF423 directly interacts with CEP290/NPHP6. (F) A human fetal brain yeast two-hybrid library screened with human CEP290/NPHP6 (JAS2; aa 1917–2479) fused to the DNA-binding domain of GAL4 (pDEST32) identified ZNF423 as a direct interaction partner of CEP290/NPHP6. The interaction was confirmed using direct yeast two-hybrid assay where 1 and 2 represent colony growth of CEP290 bait with ZNF423 prey. a–e are controls for colony growth on medium deficient in histidine, leucine and tryptophan. (G) HEK293T were cotransfected with human V5-tagged partial human V5-CEP290 clone and GFP-tagged full-length human ZNF423 clone. Immunoprecipitation with anti-V5 (lane 2), but not control IgG (lane 3) precipitated both the V5-tagged CEP290 (arrowhead) as well as GFP-tagged ZNF423 (arrow). EXPRESSION / LABELING:
|
|
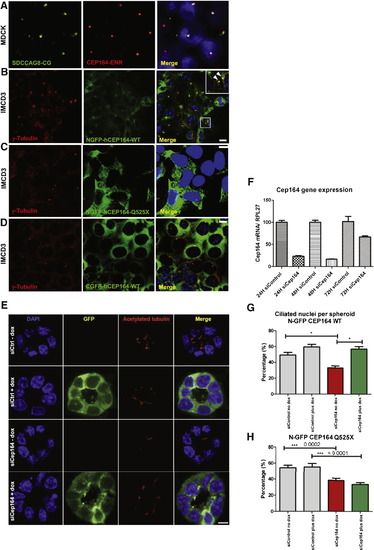
Expression of Mutant CEP164 in Renal Epithelial Cells Abrogates Localization to Centrosomes (A) Immunofluorescence using α-SDCCAG8/NPHP10-CG antibody in MDCK cells, labels both centrioles, whereas α-CEP164-ENR antibody demonstrates CEP164 staining at the mother centriole only. (B) Inducible overexpression of N terminally GFP-tagged human full-length CEP164 isoform 1 (NGFP-CEP164-WT) in IMCD3 cells demonstrates, in addition to a cytoplasmic expression pattern, localization at one of the two centrioles (inset, arrow heads) consistent with selective localization to the mother centriole Graser et al., 2007). Both centrioles are stained with an anti-γ-tubulin antibody (C) In contrast, the centrosomal signal is abrogated upon overexpression of an N terminally GFP-tagged truncated CEP164 construct representing the mutation p.Q525X. (D) The number of centrosomes positive for CEP164 is reduced upon overexpression of C terminally GFP-tagged human full-length CEP164 isoform 1 (CGFP-hCEP164-WT), which mimics the mutation p.X1460WextX57 that causes a read-through of the stop-codon X1460, adding 57 aberrant amino acid residues to the C terminus of CEP164. Similar data were obtained upon CEP164 expression in hTERT-RPE cells (see also Figures S3B–S3D). IMCD3 cells were stably transfected with the respective CEP164 constructs in a retroviral vector for doxycyclin-inducible expression (pRetroX-Tight-Pur). Scale bars, 10 μm (E–H) Knockdown of Cep164 disrupts ciliary frequency. (E) Depletion of Cep164 by siRNA (F) causes a ciliary defect in 3D spheroid growth assays. IMCD3 cells transfected with either siCtrl or siCep164 were grown to spheroids in 72 hr and immunostained for acetylated tubulin (red). DAPI stains nuclei (blue). Doxycycline induced stably transfected NGFP-hCEP164-WT (green). Space bar represents 5 μm. (G) Nuclei and cilia were scored within a single spheroid to generate ciliary frequencies. siCep164 transfected cells manifest lower cilia frequencies (33%) compared to control transfected IMCD3 cells (49%). Induction of NGFP-hCEP164-WT in siCep164 transfected cells rescues this ciliary defect (57%). 50 spheroids per condition were analyzed in three independent experiments. Error bars represent SEM, n = 3, p value < 0.0002. (H) Ciliary frequency is not rescued by mutant CEP164. Ciliary frequencies are reduced in siCep164 transfected IMCD3 cells (39%) compared to control siCtrl transfected IMCD3 cells (54%). Induction of NGFP-hCEP164-Q525X does not rescue this ciliary defect (34%). 50 spheroids per condition were analyzed. Error bars represent SEM, ***p value < 0.0002 See also Figure S3. |
|
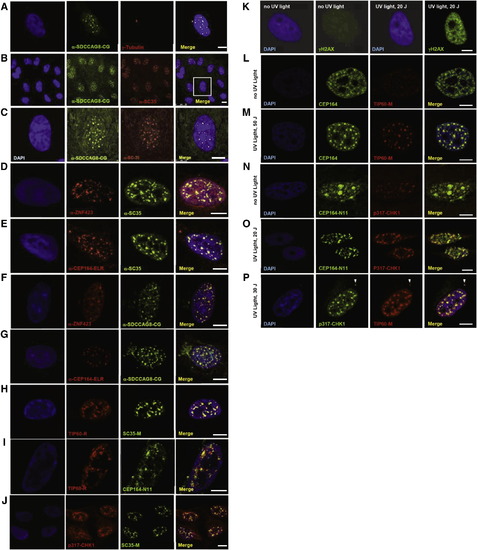
Colocalization upon Immunofluorescence of the NPHP-RC Proteins SDCCAG8/NPHP10, ZNF423 and CEP164 to Nuclear Foci that Are Positive for the DDR Signaling Proteins SC35, TIP60 and Chk1 in hTERT-RPE Cells (A–G) Colocalization of NPHP-RC proteins with SC35 in nuclear foci. SDCCAG8/NPHP10 (A-C) and ZNF423 (D) fully colocalize to nuclear foci with SC35, and (E) CEP164 partially colocalizes with SC35. SDCCAG8/ NPHP10 also colocalizes with the identified NPHP-RC proteins ZNF423 (F) and CEP164 (G). (H–J) Colocalization of NPHP-RC proteins with the DDR protein TIP60 and Chk1 to nuclear foci. (H) TIP60 fully colocalizes with SC35. (I) TIP60 partially colocalizes with CEP164. (J) Chk1 fully colocalizes with SC35/ SRSF2. DNA is stained in blue with DAPI. Scale bars, 5 μm. (K–P) Colocalization of DDR and NPHP proteins upon induction of DDR by UV radiation in HeLa cells. (K) Following irradiation of HeLa cells with UV light at 20 J/m2 a strong immunofluorescence signal of an anti-γH2AX antibody indicates activation of DDR. (L–M) Upon irradiation with UV light, CEP164-positive nuclear foci condense and colocalize with TIP60 foci of similar size. (N–O) In untreated cells (N) a pattern of broad CEP164 speckles, which are Chk1-negative and locate to DAPI-negative domains, changes to a pattern of multiple smaller foci (O) that are double positive for both CEP164-N11 and Chk1. (P) p317-Chk1 fully colocalizes with TIP60 to nuclear foci and to the centrosome (arrowhead). See also Figures S4, S5A, and S5B. |
|
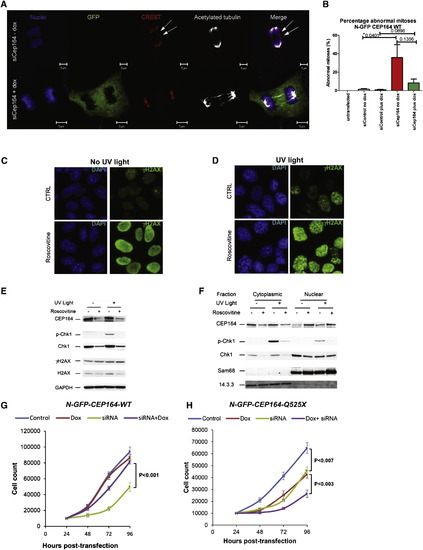
Knockdown of Cep164 Causes Anaphase Lag and Retarded Cell Growth (A and B) Knockdown of CEP164 causes anaphase lag. siCep164 knockdown in IMCD3 cells increased anaphase lag incidence from 1% after siCtrl to 21% after siCep164-treated cells (n > 250 anaphases, five independent experiments). CREST antiserum (red) and DAPI (blue) confirmed the presence of incomplete mitotic congression and unattached kinetochores during late anaphase (white arrows). Doxycycline-inducible expression of WT-CEP164 during Cep164 siRNA knockdown reduced the incidence of anaphase lag to 4%, whereas untransfected IMCD3 cells had no detectable anaphase lag (0%) (B). Bars represent SEM, p values (Student’s t test) are indicated above the bar graph. (C–F) The effect of roscovitine on UV-induced DDR. Cells were UV irradiated with 30 J/m2 and analyzed 1 hr after UV irradiation. Where indicated, cells were preincubated for 24 hr with the CDK inhibitor roscovitine (80 μM). (C and D) Immunofluorescence analysis showed that roscovitine triggered uniform nuclear distribution of γH2AX (activated H2AX phosphorylated at Ser139) in non-UV irradiated cells suggesting partial DDR activation (C). UV radiation caused enhanced γH2AX staining with a prominent nuclear foci pattern, characteristic of strong DDR activation (D). (C and D) Error bars denote SEM. (E and F) The effect of roscovitine on UV-triggered subcellular localization of CEP164 and Chk1. CEP164 and Chk1 proteins, along with nuclear marker Sam68 and cytoplasmic marker 14.3.3 were analyzed by Western blot. Roscovitine decreased the amount of CEP164 present in control and UV-irradiated cells (E). This was most likely due to translocation of CEP164 into the nucleus upon roscovitine treatment as shown by subcellular fractionation (F). As expected, UV radiation increased phosphorylation of Chk1 at Ser317 (p-Chk1) (E), and roscovitine decreased Chk1 protein expression and abrogated UV-induced p-Chk1 in both cytoplasm and nucleus (E-F). Proteins 14.3.3 and Sam68 serve as controls for cytoplasmic versus nuclear fraction, respectively. See also Figure S6. (G and H) Transient knockdown of Cep164 inhibits proliferation, which is rescued by wild-type but not mutant CEP164. In clonally selected and doxycycline (Dox)-inducible mouse IMCD3 cells siRNA knockdown was performed. (G) IMCD3 cells depleted of murine Cep164 grew more slowly (siRNA, green line) than nondepleted cells (control, blue line) or the nondepleted cells induced to express human wild-type CEP164 (Dox, red line). Expression of WT Cep164 in siRNA-depleted cells rescued the slow growth phenotype of Cep164 depletion (siRNA+dox, purple line). (H) As in (G), except mutant Cep164 cDNA (CEP164-Q525X) was expressed under doxycyclin control. Expression of this allele itself had a negative impact on cell growth (green line), suggesting a dominant negative effect. An even greater negative effect was seen when the endogenous Cep164 was depleted in cells expressing CEP164-Q525X (siRNA+dox, purple line). The average counts are plotted with standard deviations. Asterisks indicate significant differences by unpaired Student’s t test (p < 0.05). |
|
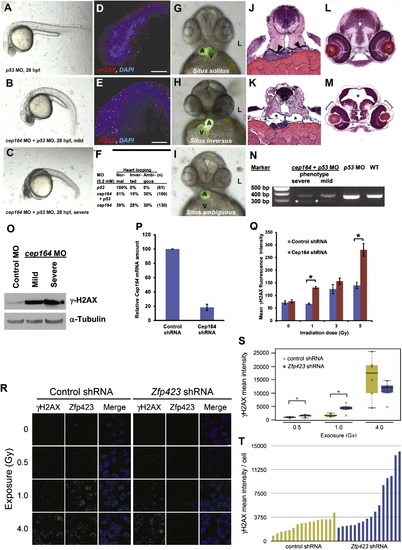
Knockdown of cep164 in Zebrafish Embryos Results in Ciliopathy Phenotypes, and Knockdown of Cep164 or Zfp423(Znf423) Causes Sensitivity to DNA Damage A morpholino-oligonucleotide (cep164 MO) targeting the exon 7 splice donor site of zebrafish cep164 was injected into fertilized eggs at the one to four-cells stage together with p53 MO (0.2 mM) to minimize nonspecific MO effects. (A–E) Whereas p53 MO injection (n = 67) did not produce any phenotype (A), coinjection of cep164 MO at 28 hpf caused the mild ciliopathy phenotype of ventral body axis curvature in 48% of embryos (60/125) (B). 50% of embryos (62/125) showed severe cell death throughout the body as judged by gray-appearing cells in the head region (C). Embryos with severe cell death also showed increased expression of phosphorylated γH2AX (D) compared to p53 MO control (E). Most embryos with massive cell death did not survive beyond 48 hpf. (F–I) At 48 hpf, surviving cep164 morphants displayed the ciliopathy phenotype of laterality defects. Whereas p53 MO did not cause any abnormal heart looping (F and G), cep164 MO caused inverted heart looping (H) or ambiguous heart looping (I). (A, atrium; L, left; V, ventricle). (J–M) At 72 hpf, cep164 morphant embryos developed further ciliopathy phenotypes. When compared to p53 MO controls (J), pronephric tubules (arrow heads) exhibited cystic dilation (K), asterisks) in 25% (7/28) of embryos, compared to p53 MO controls (J and L), 0% (0/67) of which showed kidney cysts, hydrocephalus (asterisk), or retinal dysplasia (brackets) (M). (N) At 0.2 mM, cep164 MO knockdown effectively altered mRNA processing as revealed by RT-PCR. The wild-type (WT) mRNA product is 339 bp. A shorter aberrantly spliced mRNA product appeared in cep164 morphants (asterisks), and the normal mRNA product was significantly reduced. p53 MO alone did not affect cep164 mRNA processing. (O) Quantification of γ-H2AX levels in cep164 MO morphants. Whole-fish lysates were prepared from morphants injected with control MO (p53 0.2 mM) or cep164 MO (p53 0.2 mM, cep164 0.2 mM). Injection of cep164-targeting MO causes upregulation of γ-H2AX in cep164 morphant embryos signifying perturbed DDR. γ-H2AX levels correlate with the phenotypic severity of the cep164 morphants (see A–C). Anti-α-tubulin antibody was used to show equal loading. (P and Q) Cep164-deficient IMCD3 cells exhibit radiation sensitivity. In IMCD3 cells transduced with shRNA retrovirus, Cep164 expression was suppressed by shRNA knockdown to about 20% of control as judged by qPCR (P). Cep164 knockdown resulted in a dose-dependent increase of γH2AX-positive cells in a FACS assay, signifying increased radiation sensitivity to IR and perturbed DDR. See also Figure S7. In (Q) the level of significance of two-tailed t test (p < 0.001) is indicated by an asterisk. Error bars denote SEM. (R–T) Zfp423(Znf423)-deficient P19 cells exhibit radiation sensitivity. P19 cells transduced with shRNA lentivirus were exposed to the indicated level X-irradiation. Zfp423 and γH2AX immunofluorescence was quantified in matched replicate cultures for each virus 2 hr after irradiation. (R) Representative images illustrate dose-responsiveness of γH2AX and effective knockdown of Zfp423 expression. (S) γH2AX intensity normalized to DAPI+ nuclei is increased following IR at 0.5 and 1.0 Gy, signifying increased IR sensitivity and perturbed DDR (2 fields from each of 6 replicate cultures per condition). Asterisks, uncorrected pair-wise p < 0.05, Mann-Whitney U test, 2 tails. (T) Histogram shows average γH2AX intensity per cell in 16 additional replicate cultures for each shRNA at 1.0 Gy exposure. p = 0.018, Mann-Whitney U test, 2 tails. See also Figure S7. Box plots delimit quartiles in (S). PHENOTYPE:
|
|
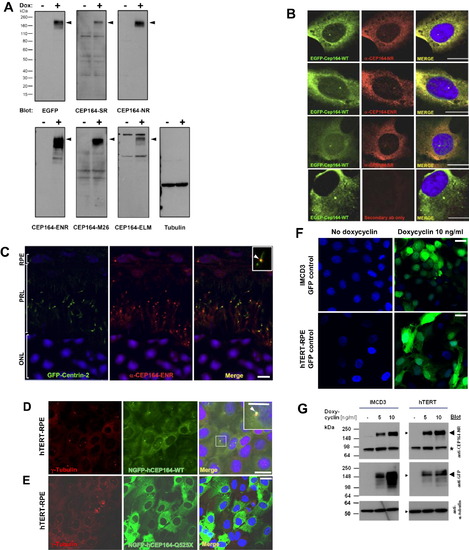
Subcellular Localization of CEP164 (A) Characterization of anti-CEP164 antibodies by immunoblotting. IMCD3 cells that doxycycline-inducibly express GFP-CEP164 isoform 1 were induced with (+) or without (-) 10 ng/ml doxycyclin for 20 hr. 20 μg of RIPA extracted cell lysates were subjected to SDS-PAGE and blotted with the indicated antibodies. Molecular weight is shown in kDa. Note that anti-CEP164 antibodies recognize a band compatible with the size of EGFP-CEP164 isoform 1 (191 kDa; arrow heads). Loading control (on right) uses an anti-αtubulin antibody. (B) Characterization of α-CEP164 antibodies by overexpression of N terminally GFP-labeled human isoform 1 wild-type construct EGFP-CEP164-WT and immunofluorescence in cell lines. Cells were costained with antibodies α-CEP164-NR, -ENR, and -SR (red), respectively, demonstrating colocalization (Merge) with N-EGFP-hCEP164-WT (green). The secondary antibody alone (lower panels) does not result in a signal. Scale bars, 5 μm. (C) CEP164 in GFP-Centrin-2 mouse photoreceptors. In photoreceptors of centrin-2-GFP mice, immunofluorescence using anti-CEP164-ENR antibody detects Cep164 at basal bodies/mother centrioles (visible as a single red dot), whereas centrin-2-GFP is expressed at both centrioles, mother and daughter. Inset shows enlargement of a representative centrosome at 3-fold higher magnification. Nuclei are stained with DAPI. Scale bars, 5 μm. ONL, outer nuclear layer; PRL, photoreceptor layer; RPE, retinal pigment epithelium. (D–G) Expression of mutant CEP164 in hTERT-RPE cells abrogates localization to centrosomes. (D) Doxycyclin (Dox)-inducible overexpression of N terminally GFP-tagged human full-length CEP164 wild-type isoform 1 (NGFP-hCEP164-WT) in hTERT-RPE (human retinal pigment epithelium) cells demonstrates, in addition to a cytoplasmic expression pattern, localization at centrosomes. (E) In contrast, the signal at centrosomes is abrogated upon overexpression of an N terminally GFP-tagged truncated CEP164 construct (NGFP-CEP164-Q525X) representing the mutation p.Q525X occurring in NPHP-RC family F59. Similar data was obtained upon CEP164 expression in IMCD3 cells (see also Figures 3B–3D). (F) Expression of GFP alone as a negative control yielded no centrosomal expression in IMCD3 or hTERT-RPE cells. hTERT-RPE cells were stably transfected with the CEP164 constructs in a retroviral vector for doxycyclin-inducible expression (pRetroX-Tight-Pur). Following selection with puromycin for 2 weeks, cells were induced with 10 ng/ml of doxycyclin for 20 hr. Cells were then costained with γ-tubulin (red) for the colocalization with NGFP-hCEP164-WT fusion proteins (green) (see also Figures 3B–3D). Scale bars, 10 μm. (G) Immunoblot of IMCD3 and hTERT-RPE cells expressing inducible GFP-CEP164 constructs confirmed doxycyclin-inducible NGFP-CEP164 fusion protein expression (<191 kDa) both with α-CEP164-NR and α-GFP antibody blotting (arrow heads). The band at <85 kDa (asterisk) may represent the endogenous isoform 2 of CEP164 (84 kDa). IMCD3 and hTERT-RPE cells were stably transfected with the N terminally tagged-GFP-CEP164 construct in a retroviral vector (pRetroX-Tight-Pur) for doxycyclin-inducible expression. Cells were selected on puromycin for 2 weeks. Both cell lines were plated and treated with the indicated doses of doxycycline and lyzed with RIPA buffer 24 hr after induction. A total of 50 μg of cell lysates were loaded onto SDS-PAGE and blotted with the antibodies indicated. The same membranes were reprobed with anti-α-tubulin antibody as an indicator of equal loading (GFP-blotted membrane shown) (see also Figure 3). See also Figure 3. |
|
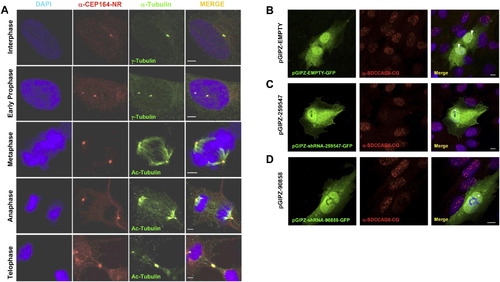
Subcellular Localization of CEP164 and SDCCAG8 by Immunofluorescence (A) CEP164 colocalizes with the mother centriole (labeled with γ-tubulin), the mitotic spindle poles (actetylated tubulin), and the midbody throughout the cell cycle in hTERT-RPE cells. Whereas in the centriole-engaged state (upper panel) γ-tubulin antibody labels both centrioles, the α-CEP164 antibody only labels one centriole (the mother centriole) in hTERT cells. Immunofluorescence of endogenous CEP164 was performed in hTERT cells. Cells were fixed (4% PFA), permeabilized (0.1% SDS) and immuno-stained with antibody anti-CEP164-NR (red) and costained with anti-γ-tubulin or anti-acetylated tubulin antibodies (green). DAPI was used to label DNA (Blue). Scale bars, 2.5 μm. (B)–(D) Upon immunofluorescence (IF) in hTERT-RPE cells antibody α-SDCCAG8-CG recognizes nuclear foci that are absent upon transient SDCCAG8/NPHP10 knockdown. Left panels show transfected hTERT-RPE cells, middle panels show IF with α-SDCCAG8-CG, right panels show merge of left and middle panels. Antibody α-SDCCAG8-CG recognizes nuclear foci in hTERT-RPE cells (middle and right panels). (B) hTERT cells transiently transfected with GFP-labeled negative control shRNA construct exhibit nuclear foci upon IF with α-SDCCAG8-CG (arrow heads in right panel), whereas in cells transfected with GFP-labeled SDCCAG8 shRNA knockdown constructs pGIPZ-259547 (C) or pGIPZ-90858 (D) nuclear foci are absent (asterisks in right panels of (C) and (D), demonstrating specificity of the nuclear foci signal detected by α-SDCCAG8-CG. Scale bars, 5 μm. See also Figure 4. |
|
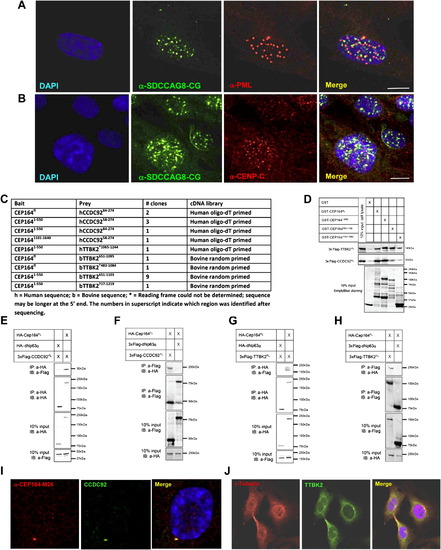
(A–B) Immuofluorescence Imaging of the NPHP-RC Protein SDCCAG8/NPHP10 with Other Proteins of Nuclear Foci in hTERT-RPE Cells, and (C–J) Identification of CCDC92 and TTKB2 as Direct Interaction Partners of CEP164 (A–B) SDCCAG8 labeled with antibody α-SDCCAG8-CG exihibits a similar number and size of nuclear foci in comparison to signals from antibodies against the nuclear foci markers promyelocytic leukemia protein (PML) (A) and centromere protein C (CENP-C) (B). However, these proteins do not colocalize with SDCCAG8. Scale bars, 5 μm. (C–J) Identification of CCDC92 and TTKB2 as direct interaction partners of CEP164. (C) Four baits, BD-CEP164fl, BD-CEP1641-550, BD-CEP164551-1100, and BD-CEP1641101-1640 were used to screen two retinal cDNA libraries, a human oligo-dT primed library and a bovine randomly primed library, employing a GAL-4 based yeast two-hybrid system. CEP164 was found to interact with two proteins, coiled coil domain containing protein 92 (CCDC92) and tau tubulin kinase 2 (TTBK2). The CEP164, CCDC92 and TTBK2 fragments are indicated with amino acids numbering in superscript. (D) Lysates from COS-1 cells transfected with 3xFlag-CCDC92fl or 3xFlag-TTBK2fl were used in a pull-down assay of GST fusion proteins GST-CEP164fl (191kDa), GST-CEP1-550 (87kDa), GST-CEP164551-1100 (92kDa), and GST-CEP1641101-1640 (68kDa). 3xFlag-CCDC92fl preferentially binds GST-CEP164fl and GST-CEP1641-550, while 3xFlag-TTBK2fl binds all four GST fusion proteins. The results are further confirmed by unbound GST (26kDa), which shows no interaction with 3xFlag-CCDC92fl nor 3xFlag-TTBK2fl. (E–F) For coimmunoprecipitation 3xHA-CEP164fl in combination with 3xFlag-CCDC92fl were overexpressed in COS-1 cells (10% protein input shown). In an anti-HA immunoprecipitation 3xHA-CEP164fl coprecipitated with 3xFlag-CCDC92fl (E), whereas the coprecipitation with the unrelated 3xHA-dNp63 is very limited. This was confirmed in the reciprocal coIP using an anti-Flag antibody against 3xFlag-CCDC92fl (F). (G–H) For coimmunoprecipitation 3xHA-CEP164fl in combination with 3xFlag-TTBK2fl were overexpressed in COS-1 cells (10% protein input shown). In an anti-HA immunoprecipitation of 3xHA-CEP164fl, this protein coprecipitated with 3xFlag-TTBK2fl (G), whereas the coprecipitation with the unrelated 3xHA-dNp63 was negative. This was confirmed in the reciprocal coIP using an anti-Flag antibody to immunoprecipitate 3xFlag-TTBK2fl (H). (I–J) (I) The α-CCDC92 antibody signal fully colocalizes with α-CEP164-M26 at the mother centriole upon immunofluorescence imaging in hTERT-RPE cells. (J) TTBK2 weakly colocalizes with α-CEP164-M26 at one of the centrioles, but yields a strong signal at the mid body in dividing hTERT-RPE cells. See also Figure 4. |
|
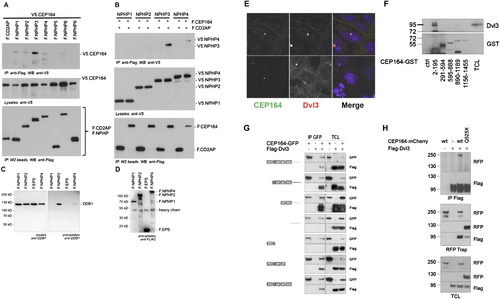
Molecular Interaction of NPHP-RC Gene Products with DDR Proteins. (A and B) Interaction of NPHP3 with CEP164. (A) To determine whether CEP164 interacts with known NPHP proteins, HEK293T cells were cotransfected with expression constructs for N terminally V5-tagged human full-length CEP164 and FLAG-tagged human full length NPHP1-NPHP5, NPHP8, NPHP9, or the control protein CD2AP as indicated. Comparable amounts of CEP164 were present in all lysates (middle panel). NPHP proteins were precipitated, using anti-Flag M2 beads (bottom panel). NPHP3 immobilized V5-tagged CEP164; a weak interaction was also detectable for NPHP1, 2 and 4 (top panel). (B) The reciprocal interaction confirmed an interaction between CEP164 and NPHP3. Flag-tagged CEP164 or CD2AP were coexpressed with V5-tagged NPHP1-NPHP4. Precipitation of NPHP3, but not the control protein CD2AP immobilized V5-tagged NPHP3. A weak, but reproducible interaction was also detected for NPHP4 (top panel). (C and D) Interaction of NPHP2 with DDB1. (C) HEK293T cells were transiently transfected with the Flag-tagged NPH-proteins NPHP1, NPHP2, NPHP4 or a control protein (Flag-EPS). Following immunoprecipitation with Flag-antibody Western blot analysis revealed that endogenous DDB1 coprecipitated with NPHP2 and to a lesser extent with NPHP4 but not with NPHP1 or the control protein. (D) Control experiment for protein expression using anti-FLAG antibody. (E and H) Cep164 and Dvl3 are in a precipitable complex. (E) CEP164 and Dvl colocalize at centrosomes. Whereas CEP164 is known to localize to the mother centriole only, Dvl3 is noted to label both centrioles (upper panel) or to the daughter centriole only (lower panel). HEK293 cells were fixed, labeled with antibody α-CEP164-PR and α-Dvl3. (F) Domain mapping of the CEP164 interaction with Dvl3. Series of bacterially produced CEP164-GST fragments were incubated with HEK293 cell lysates, precipitated using GST pull-down, and probed for presence of Dvl3 by immunoblotting against Dvl3 and GST, respectively. Endogenous Dvl3 interacts with a region within the first 194 amino acids of CEP164, which corresponds to the ATRIP binding domain. (G) CEP164 interacts with the proline-rich region of Dvl3. HEK293 cells were transfected with GFP-tagged full-length human CEP164 and a panel of truncation mutants of Flag-Dvl3 as indicated, and coimmunoprecipitation was performed with an anti-GFP antibody. Note that CEP164 only interacts with Dvl3 fragments that contain the proline-rich region (Pro). (H) Full length CEP164 but not the mutant CEP164-Q525X interacts with Dvl3. HEK293 cells were transfected with the indicated plasmids and subjected to immunoprecipitation using antibodies against FLAG-Dvl3 (upper panel) or CEP164-mCherryRFP (middle panel). Immunoblot analysis demonstrates strong interaction with Dvl3 for wild-type (wt) CEP164-mCherryRFP, whereas interactions is strongly reduced for CEP164-Q525X to Dvl3. |
|
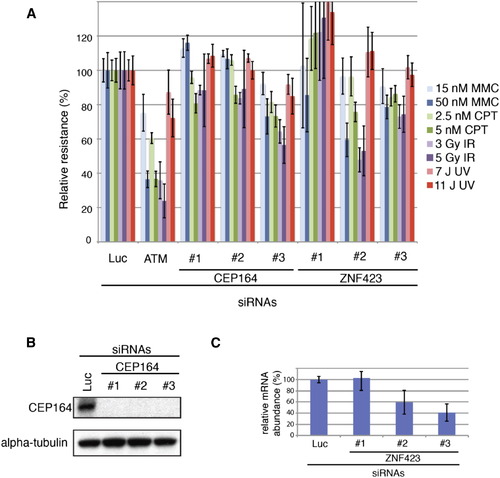
Depletion of CEP164 or ZNF423(Zpf423) Results in Sensitivity to DNA Damaging Agents (A) Multicolor competition assay (MCA) (Smogorzewska et al., 2007). GFP-U2OS cells were transfected with the indicated siRNAs against CEP164 and ZNF423. Seventy-two hours after after transfection they were mixed with RFP-U2OS cells transfected with Luciferase (Luc) and DNA damage was induced using the indicated agents. After 7 days, percent of gfp-positive cells was measured using fluorescence-activated cell sorting and the resistance to DNA damage was calculated in comparison to untreated cells. DNA damage resistance was set at 100% for GFP-U2OS cells transfected with an siRNA against Luciferase (Luc). Cells transfected with siRNAs against ATM were used as a positive control. Experiments were done in triplicate. Standard deviations are indicated. Depletion of CEP164 (B) led to sensitivity to camptothecin (CPT) and gamma irradiation (IR). In addition, depletion with siRNA #3 led to sensitivity to mitomycin C (MMC). Furthermore, transfection of two separate ZNF423 siRNAs (#2 and #3), that resulted in ZNF423 mRNA depletion as assessed by RT-qPCR (C), caused sensitivity to MMC, CPT, and IR (L). An siRNA that did not result in ZNF423 mRNA depletion (C), siRNA #1 did not result in increased sensitivity to DNA damage. (B) Western blot analysis of CEP164 expression after siRNA transfection in cells used in the MCA assay shown in (A) (C) RT-qPCR in U2OS cells transfected with the three siRNAs against ZNF423 used in the MCA assay shown in (A). Reactions were performed in triplicate. Standard deviations are indicated. See also Figure 6. |
Reprinted from Cell, 150(3), Chaki, M., Airik, R., Ghosh, A.K., Giles, R.H., Chen, R., Slaats, G.G., Wang, H., Hurd, T.W., Zhou, W., Cluckey, A., Gee, H.Y., Ramaswami, G., Hong, C.J., Hamilton, B.A., Cervenka, I., Ganji, R.S., Bryja, V., Arts, H.H., van Reeuwijk, J., Oud, M.M., Letteboer, S.J., Roepman, R., Husson, H., Ibraghimov-Beskrovnaya, O., Yasunaga, T., Walz, G., Eley, L., Sayer, J.A., Schermer, B., Liebau, M.C., Benzing, T., Le Corre, S., Drummond, I., Janssen, S., Allen, S.J., Natarajan, S., O'Toole, J.F., Attanasio, M., Saunier, S., Antignac, C., Koenekoop, R.K., Ren, H., Lopez, I., Nayir, A., Stoetzel, C., Dollfus, H., Massoudi, R., Gleeson, J.G., Andreoli, S.P., Doherty, D.G., Lindstrad, A., Golzio, C., Katsanis, N., Pape, L., Abboud, E.B., Al-Rajhi, A.A., Lewis, R.A., Omran, H., Lee, E.Y., Wang, S., Sekiguchi, J.M., Saunders, R., Johnson, C.A., Garner, E., Vanselow, K., Andersen, J.S., Shlomai, J., Nurnberg, G., Nurnberg, P., Levy, S., Smogorzewska, A., Otto, E.A., and Hildebrandt, F., Exome Capture Reveals ZNF423 and CEP164 Mutations, Linking Renal Ciliopathies to DNA Damage Response Signaling, 533-548, Copyright (2012) with permission from Elsevier. Full text @ Cell