- Title
-
Neuron-specific expression of atp6v0c2 in zebrafish CNS
- Authors
- Chung, A.Y., Kim, M.J., Kim, D., Bang, S., Hwang, S.W., Lim, C.S., Lee, S., Park, H.C., and Huh, T.L.
- Source
- Full text @ Dev. Dyn.
|
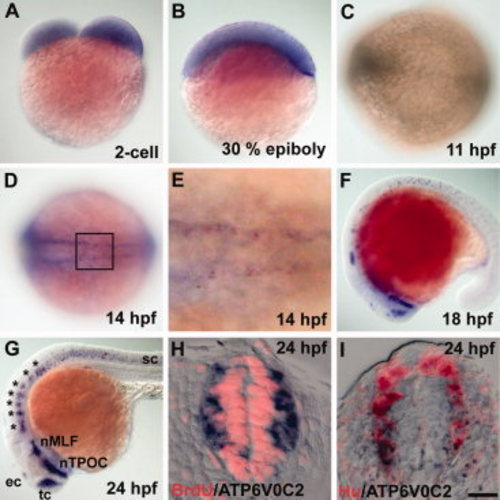
atp6v0c2 expression revealed by in situ RNA hybridization. A, B: Lateral views of (A) 2-cell stage and (B) 30% epiboly-stage embryos, dorsal at the top. C, D: Dorsal view of the trunk region of 11-hpf (C) and 14-hpf (D) embryos, anterior to the left. E: High-magnification image of the boxed area in D. F, G: Lateral views of 18-hpf (F) and 24-hpf (G) embryos, anterior to the left. tc, telencephalic cluster; ec, epiphysial cluster; nTPOC, nucleus of the tract of the post optic commissare; nMLE, nucleus of the medial longitudinal fasciculus; sc, spinal cord. H, I: Transverse sections of the spinal cord of a 24-hpf embryo, dorsal at the top. H: Sections were processed by in situ hybridization with an atp6v0c2 RNA probe and subsequent labeled with anti-BrdU antibody (red staining). I: Sections were processed by in situ hybridization with an atp6v0c2 RNA probe and subsequent labeling with anti-Hu antibody (red staining). Scale bar: A–D, F, G, 80 μm; E, H, I, 20 μm. EXPRESSION / LABELING:
|
|
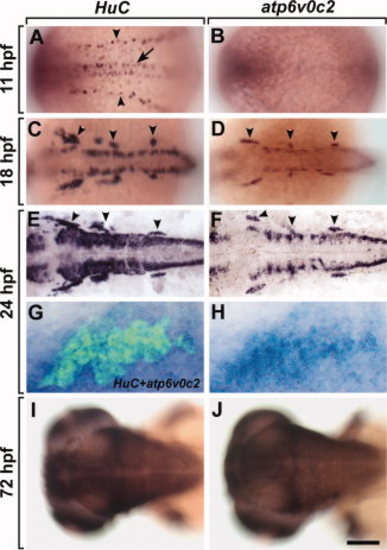
Spatial and temporal expression patterns of atp6v0c2 and HuC analyzed by in situ RNA hybridization. A–J: Dorsal views, anterior to the left, of whole embryos. A: Expression of HuC in primary motor neurons (arrow) and Rohon-Beard cells (arrowheads). B: There is no detectable expression of atp6v0c2 in 3-somite stage (11-hpf) embryos. Expression of HuC(C, E) and atp6v0c2(D, F) in 18-hpf embryos (C, D) and 24-hpf embryos (E, F). Arrowheads indicate cranial ganglions. G, H: Expression of atp6v0c2 RNA alone (H) and subsequent labeling of the same embryo with anti-Hu antibody (G) (green fluorescence) at the trigeminal ganglion of the 24 hpf embryo. I, J: Expression of HuC (I) and atp6v0c2(J) in the brain of 72-hpf embryos. Scale bar: A–F, I, J, 60 μm; G, H, 10 μm. EXPRESSION / LABELING:
|
|
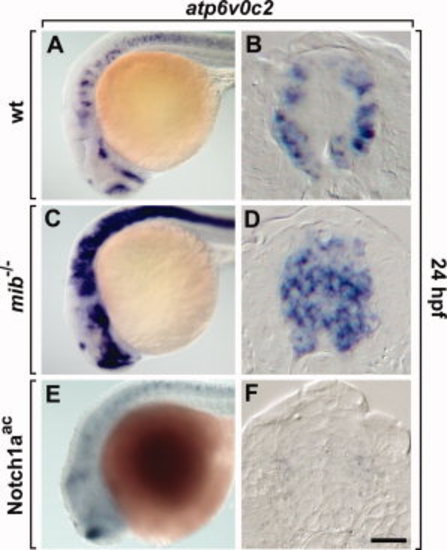
Manipulation of Notch activity results in alteration of atp6v0c2 expression in the zebrafish CNS. A, C, E: Lateral views of the whole embryo brain, anterior to the left. B, D, F: Transverse sections of the spinal cord, dorsal to the top. Expression of atp6v0c2 in wild-type (A, B), mib-/- mutant (C, D), and Notch1aac transgenic embryos induced by heat-shock (E, F). Scale bar: A, C, E, 80 μm; B, D, F, 20 μm. EXPRESSION / LABELING:
|
|
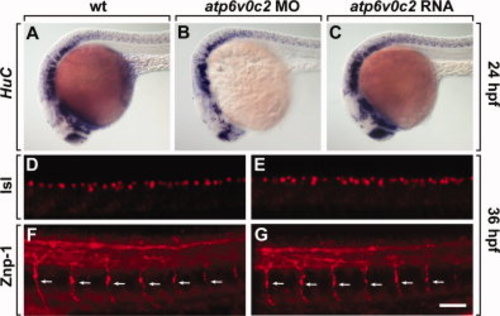
ATP6V0C2 function is dispensable for neurogenesis. All panels show lateral views of whole embryos, anterior to the left. A–C: Wild-type (A), atp6v0c2 MO-injected (B), and atp6v0c2 RNA-injected (C) embryos analyzed by HuC RNA in situ hybridization. D, E: Labeling with an anti-Isl antibody to stain for sensory neurons in the dorsal spinal cord of wild-type (D) and atp6v0c2 MO-injected (E) embryos. F, G: Labeling with an anti-Znp-1 antibody to stain for motor axon fibers in the peripheral nervous system of wild-type (F) and atp6v0c2 MO-injected (G) embryos. Arrows indicate motor axon fibers. Scale bar: A–F, 100 μm; G–J, 40 μm. EXPRESSION / LABELING:
|
|
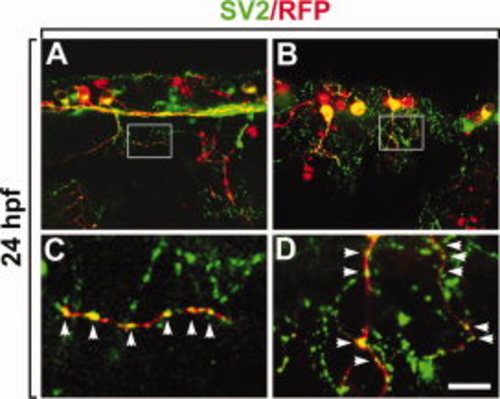
ATP6V0C2 proteins are localized in the presynaptic vesicles. All panels show lateral views of the spinal cord, anterior to the left. A, B: Wild-type embryos injected with HuC:apt6v0c2-rfp DNA (red fluorescence) and labeled with an anti-SV2 antibody to detect presynaptic vesicles (green fluorescence). C, D: High-magnification images of the boxed areas in A and B, respectively. Scale bar: A, B, 20 μm; C, D, 4 μm. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|
|
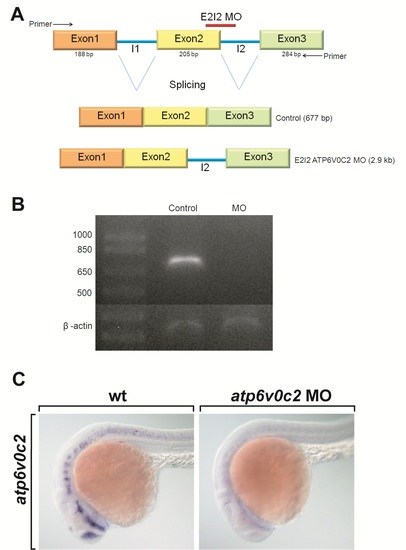
Knock-down of atp6v0c2 in zebrafish embryos. A: Schematic diagrams of atp6v0c2 genomic DNA and splice sites targeted by E2I2 MOs (atp6v0c2 MO). atp6v0c2 MOs block the excision of the intron between exon 2 and exon 3, generating abnormal atp6v0c2 mRNA in which exon 2 and exon 3 are separated by the second intron (I2). B: Confirmation of the efficiency of atp6v0c2 MOs by reverse transcriptase-PCR (RT-PCR) analysis. RT-PCR with RNA from control embryos amplifies a 677-bp segment of the transcript. However, RT-PCR using RNA from atp6v0c2 MO-injected embryos failed to generate a PCR product, because excision of the 2,169-bp intron (I2) between exon 2 and exon 3 did not occur. C: Confirmation of the efficiency of atp6v0c2 MOs by in situ hybridization with atp6v0c2 RNA probe. All panels show lateral views of wild-type and atp6v0c2 MO-injected embryos analyzed by atp6v0c2. Since atp6v0c2 MOs interfere with splicing between exon2 and exon3 and generate an mRNA split by intron2, whole-mount RNA in situ hybridization did not detect any atp6v0c2 expression in the embryos injected with atp6v0c2 MOs. |