|
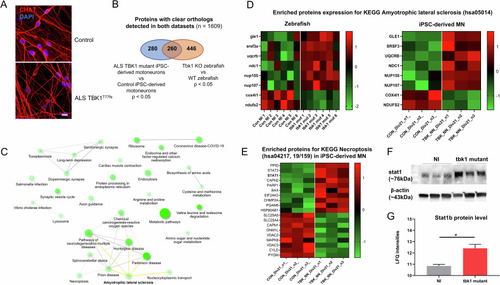
Joint proteomic analysis of tbk1 mutants zebrafish and iMNs from ALS patients with TBK1 mutations. A Fluorescent immunolabeling of the neuronal marker CHAT in differentiated induced pluripotent stem cell-derived motor neurons (iMNs) at day 21 from an ALS patient harboring the loss of function mutation TBK1T77fs compared to control iMNs. B Proteomic profiles of ALS-TBK1 iMNs compared to control iMNs. The differentially expressed protein list was compared to the list of zebrafish orthologs differentially expressed in tbk1-deficient zebrafish larvae, and enrichment analysis was performed on the common list of 260 terms. C Network plot showing the relationship between enriched pathways from TBK1-mutant ALS models. A close interaction between the necroptosis pathway and several neurodegenerative pathways, including the amyotrophic lateral sclerosis (ALS) KEGG pathway, is observed. D Heatmap representation of protein levels (row z scores) for significantly enriched proteins in both datasets associated with the KEGG term ALS in tbk1 mutant vs NI zebrafish and ALS-TBK1 iMNs vs control iMNs. E Heatmap representation of protein levels (row z scores) for significantly enriched proteins associated with the KEGG term necroptosis in ALS-TBK1 iMNs vs control iMNs. F Western blotting of STAT1 expression in tbk1 mutant and NI zebrafish. G Mass spectrometry quantification of stat1b protein level in tbk1 mutant and NI zebrafish.
|