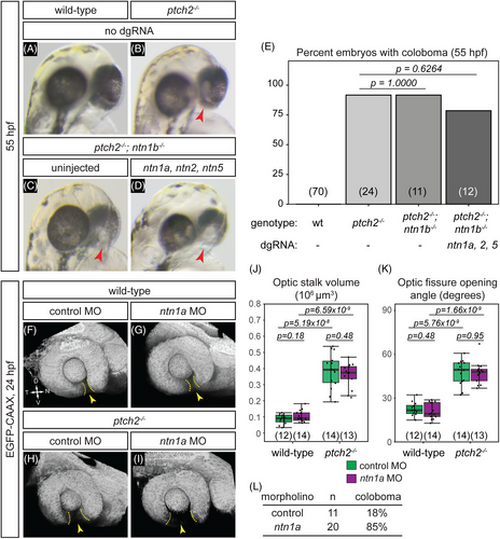
Netrin ligands are not required for the ptch2−/− coloboma phenotype. (A-E) Loss of netrin genes using CRISPR mutagenesis does not rescue the ptch2−/− coloboma phenotype. All embryos were evaluated for coloboma, genotyped, and gRNA-injected embryos were then individually quantified for mutagenesis efficiency. For gRNA-injected embryos, only ptch2 −/−;ntn1b−/− embryos with >70% alleles mutated for the remaining three genes were analyzed. (A) wild-type (wt), (B) ptch2−/−, (C) ptch2−/−; ntn1b−/−, (D) ptch2−/−; ntn1b−/− injected with ntn1a, ntn2, and ntn5 dgRNA, 55 hpf. Red arrowheads, coloboma. (E) Percentage of embryos with coloboma. Numbers in parentheses at base of graph indicate n. (–L) Morpholino-mediated knockdown of ntn1a does not rescue the ptch2−/− optic fissure and stalk phenotypes. (F) wild-type injected with control MO; (G) wild-type injected with ntn1a MO; (H) ptch2−/− injected with control MO; (I) ptch2−/− injected with ntn1a MO. Tissue morphology visualized with Tg(bactin2:EGFP-CAAX) transgene (grayscale), 24 hpf. Yellow dotted lines, optic fissure margins; yellow arrowheads, optic fissure opening. (J, K) Quantification of optic stalk volume (J) and optic fissure opening angle (K) at 24 hpf, neither of which is affected by ntn1a MO injection. (L) Quantification of coloboma in embryos from ptch2+/−; Tg(bactin2:EGFP-CAAX) incross injected with the control MO, or the ntn1a MO. Knockdown of ntn1a in the ptch2−/− results in a coloboma phenotype in 85% of embryos. n = total number of embryos screened.
|

