Fig. 2
- ID
- ZDB-FIG-240219-10
- Publication
- Murali et al., 2024 - Genetic variant classification by predicted protein structure: A case study on IRF6
- Other Figures
- All Figure Page
- Back to All Figure Page
|
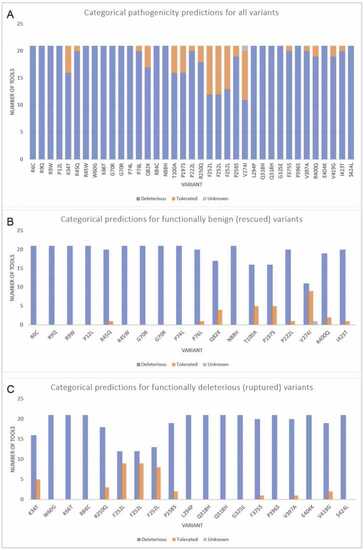
Pathogenicity predictions of IRF6 variants from categorical tools (A) This graph shows the distribution of predictions for each variant from tools that give a categorical prediction (ex. deleterious or benign). Based on these results, computational tools are more likely to predict variants to be deleterious. Interestingly, the nucleotide variants that resulted in an amino acid change in F252 showed varying results, even though all 3 variants result in the same amino acid change. One tool (M-CAP) did not provide a prediction for V274I and is thus marked as unknown. Out of the 37 variants, 19 variants were unanimously predicted to be deleterious by all computational tools. Graphs B and C depict the breakdown of predicted clinical significance from categorical computational tools for (B) variants that ‘rescued’ the irf6 -/- zebrafish (benign) and (C) variants that remained as ‘ruptured’ (deleterious). |