Fig. 1
- ID
- ZDB-FIG-231229-7
- Publication
- Nikaido et al., 2023 - Intestinal expression patterns of transcription factors and markers for interstitial cells in the larval zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
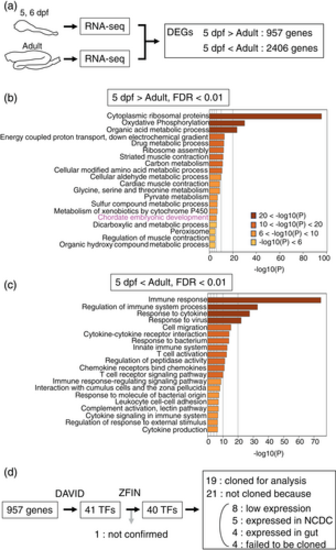
Identification of transcription factors expressed more in larval gut. (a) A schematic drawing of processes of identification and expression analysis of transcription factors (TF) in this study. Differentially expressed genes (DEGs) are identified by comparing RNA-seq results between adult and larval (5, 6 dpf) gut. Identified TFs are subjected to analysis of expression patterns by in situ hybridization. (b) Enrichment analysis of DEGs expressed more in larval gut by using 957 genes with FDR values below 0.01. Genes related with “Chordate embryonic development” are enriched in this gene group. (c) Similar enrichment analysis of DEGs expressed more in adult gut by using 2,406 genes. Genes related with, for example, “Immune response” and “Regulation of immune system process” are enriched most in this gene group. Log10(P): Ordinary logarithm of P(robability)-value. (d) Selection of 19 genes for cloning from 957 genes enriched in larval gut. The “Gene functional classification” provided by DAVID predicted 41 genes as transcription factors. Among them, 40 genes were also predicted to have DNA-binding activity in ZFIN. Because one gene, gastrula zinc finger protein XlCGF71.1-like (Entrez ID 103909501), was not found in ZFIN, we exclude it for further analysis. Among the 40 genes, 19 genes were cloned, and the remaining were not because of the reasons described. |