Fig 3
- ID
- ZDB-FIG-231024-25
- Publication
- Emmerich et al., 2023 - Transcriptomic comparison of two selective retinal cell ablation paradigms in zebrafish reveals shared and cell-specific regenerative responses
- Other Figures
- All Figure Page
- Back to All Figure Page
|
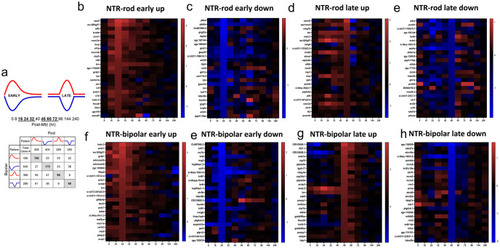
Identification of early and late DEG patterns and heatmaps for top DEGs in each. (a) DEGs for each paradigm were split into upregulated (red) and downregulated (blue) as well as early (16-32h) and late (48-72h) based on peaks in |