Figure 5.
- ID
- ZDB-FIG-231024-12
- Publication
- Massaquoi et al., 2023 - Cell-type-specific responses to the microbiota across all tissues of the larval zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
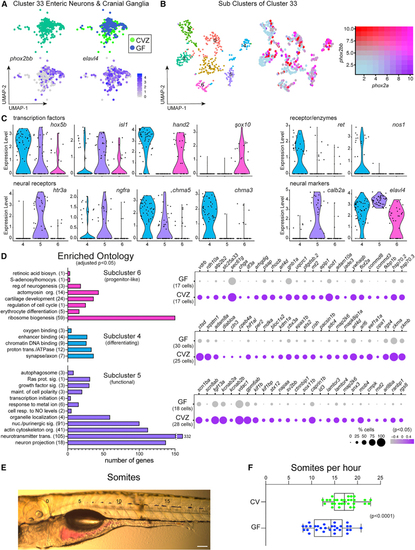
Enteric neurons are transcriptionally heterogeneous in response to the microbiota (A) Cluster 33 includes enteric neurons and cranial ganglia showing expression of (B) uMAP plots show transcriptional heterogeneity of cluster 33 with a range in co-expression of (C) Violin plots compare expression of enteric neuron transcription factors, neural receptors, and neural markers within subclusters 4, 5, and 6. (D) GO analysis plots represent genes enriched within subcluster 4, 5 or 6 relative to the other subclusters. The corresponding dot plots represents differential gene expression in CVZ versus GF cells for each subcluster. (E) Image displays a larval zebrafish post-gavage with phenol red dye. Numbers labeled on the image indicate the somite number. Scale bar of 100 μm corresponds to the micrograph. (F) Horizontal boxplots represent the rate of transit for the red dye post-gavage with respect to somite distance. In the box plots, the middle line representes the median, the right and left sides of the box represent the upper and lower quartiles, the whiskers represent the min and max values, and the plus symbol represents the mean. Each dot symbol represents data from a single 6 dpf zebrafish. The p value displayed is a result from Student’s t test. |