Figure 3
- ID
- ZDB-FIG-220507-25
- Publication
- Thumberger et al., 2022 - Boosting targeted genome editing using the hei-tag
- Other Figures
- All Figure Page
- Back to All Figure Page
|
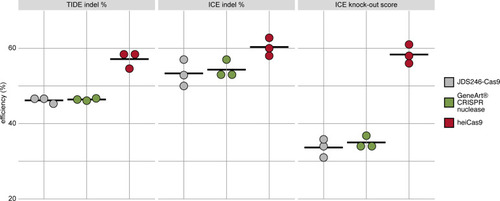
Mouse SW10 cells were co-transfected with MmPrx crRNA and mRNAs of JDS246-Cas9, GeneArt CRISPR nuclease, and heiCas9, respectively. Genome editing efficiency was assessed by Tracking of Indels by Decomposition (TIDE) and Inference of CRISPR Editing (ICE) tools. ICE knock-out score represents proportion of indels that indicate a frameshift or≥21bp deletion. Data points represent three biological replicates, black line indicates respective mean: TIDE indel %: JDS246-Cas9 = 46.2; GeneArt CRISPR nuclease = 46.4, heiCas9 = 57.1; ICE indel %: JDS246-Cas9 = 53.3; GeneArt CRISPR nuclease = 54.3, heiCas9 = 60.3; ICE knock-out score %: JDS246-Cas9 = 33.7; GeneArt CRISPR nuclease = 35.0, heiCas9 = 58.3. R2> 0.9 (TIDE) and>0.9 (ICE) for all mRNAs tested. For representative indel spectrum for each mRNA, see Figure 3—figure supplement 1.
|