Figure 6—figure supplement 1.
- ID
- ZDB-FIG-220326-38
- Publication
- Ahmed et al., 2022 - Strip1 regulates retinal ganglion cell survival by suppressing Jun-mediated apoptosis to promote retinal neural circuit formation
- Other Figures
-
- Figure 1
- Figure 1—figure supplement 1
- Figure 1—figure supplement 2.
- Figure 2
- Figure 2—figure supplement 1
- Figure 2—figure supplement 2.
- Figure 3
- Figure 3—figure supplement 1—source data 1.
- Figure 3—figure supplement 2.
- Figure 4
- Figure 4—figure supplement 1.
- Figure 5
- Figure 5—figure supplement 1.
- Figure 5—figure supplement 2
- Figure 6
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 2—source data 1.
- Figure 7
- Figure 8.
- All Figure Page
- Back to All Figure Page
|
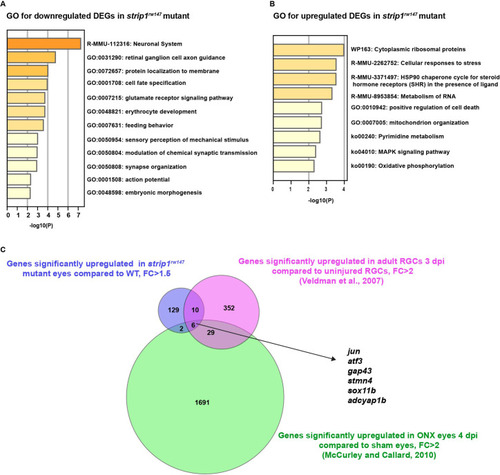
List of enriched GO terms in DEGs downregulated ( |