Fig. 3
- ID
- ZDB-FIG-210228-9
- Publication
- Duan et al., 2021 - Chromatin architecture reveals cell type-specific target genes for kidney disease risk variants
- Other Figures
- All Figure Page
- Back to All Figure Page
|
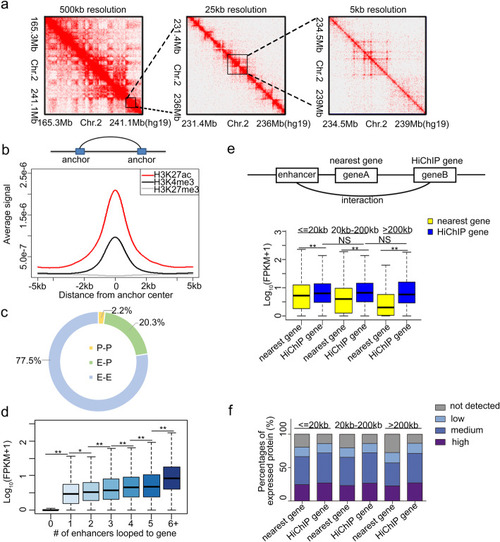
Chromatin interaction map in tubular cells. |