- Title
-
Chromatin architecture reveals cell type-specific target genes for kidney disease risk variants
- Authors
- Duan, A., Wang, H., Zhu, Y., Wang, Q., Zhang, J., Hou, Q., Xing, Y., Shi, J., Hou, J., Qin, Z., Chen, Z., Liu, Z., Yang, J.
- Source
- Full text @ BMC Biol.
|
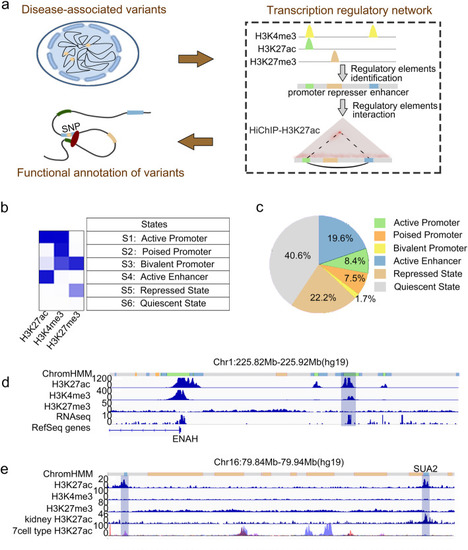
Identification of regulatory elements in tubular cells. |
|
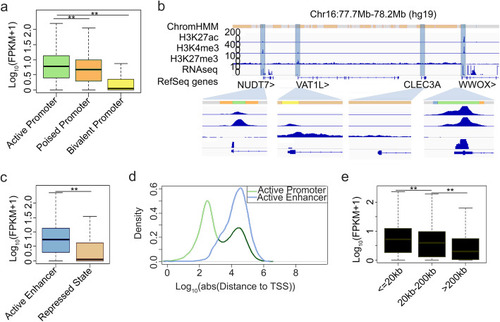
Effect of regulatory elements on gene transcription regulation. |
|
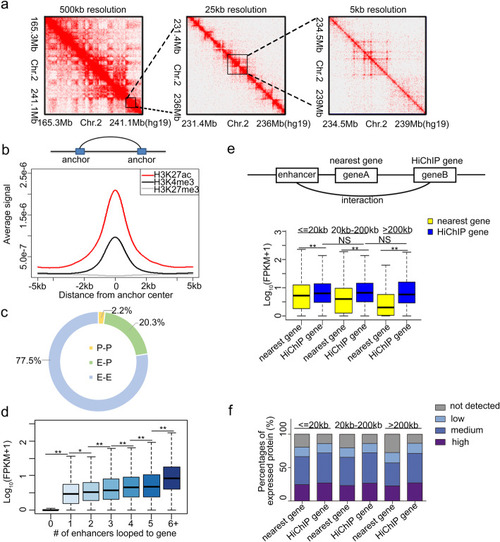
Chromatin interaction map in tubular cells. |
|
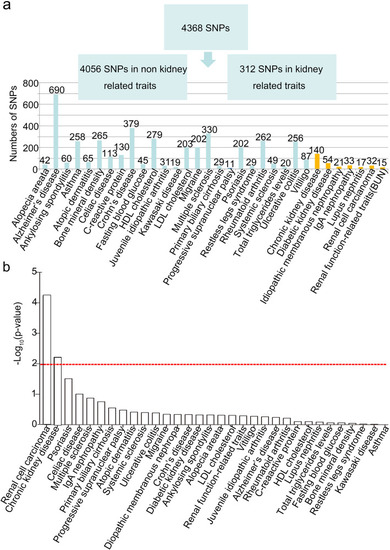
Discovery of functional SNPs for kidney related traits. |
|
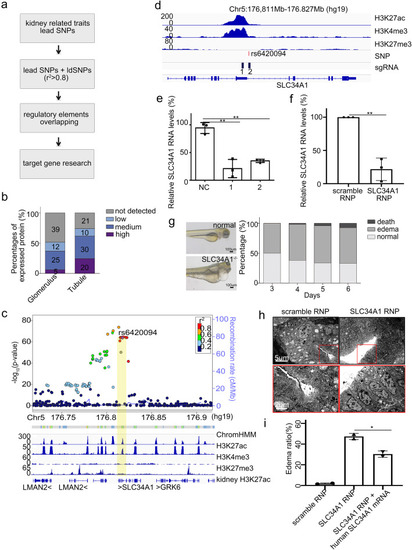
Identification of target genes for trait-associated SNPs in promoters. a Flowchart for the identification of target genes for kidney related traits. b The expression of target genes for SNPs overlapping with promoters in glomeruli and tubules. The expression categories are characterized based on immunochemistry results by Human Protein Atlas (HPA). The numbers represent the number of genes in each category. c Top: LocusZoom plot from CKD GWAS around the region of rs6420094. Bottom: epigenomic profile including ChromHMM, histone modifications in tubular cells, histone modification in kidney, and gene annotation track. Yellow bar highlights rs6420094. d Tracks of histone modifications, locations of risk SNP rs6420094 and sgRNAs designed to inactivate the element. e qPCR of SLC34A1 transcription with or without CRISPRi of the element. ** represents p value < 0.01. Each group was repeated three times. f qPCR of SLC34A1 transcription in zebrafish with RNP (sgRNA and CRISPR/Cas9 protein complex) of scramble, SLC34A1 sgRNA. Each group was repeated three times. g Left: representative figures show edema of SLC34A1−/− zebrafish. Right: edema ratio of SLC34A1−/− zebrafish at different days. h Electron microscope of kidney from zebrafish with scramble, SLC34A1 RNP. The red boxes (top) show increase space between tubules, and they are enlarged (bottom) to show the shed off of brush-border fragment. i Overexpression of human SLC34A1 mRNA partially rescue the edema. Each group were examined on two batches |
|
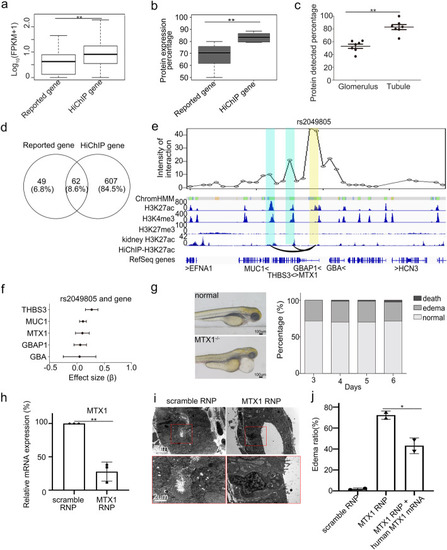
Identification of target genes for trait-associated SNPs overlapping with enhancers. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |