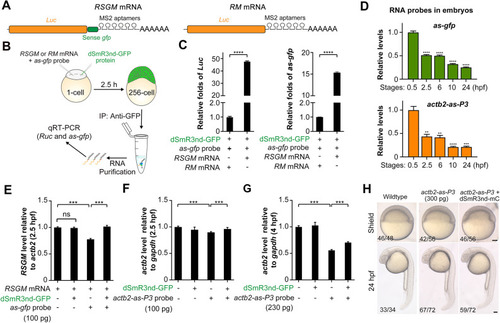
In vivo binding of dSmR3 with dsRNA and effect on target mRNA stability in the zebrafish embryo. (A) Illustration of RSGM and RM mRNA compositions. (B) Scheme of RNA immunoprecipitation: 300 pg RSGM or RM mRNA plus 350 pg as-gfp probe as well as 1 ng dSmR3nd-GFP protein were sequentially injected into one-cell-stage embryos. The dose was the amount per embryo. Approximately 2100 embryos in each group were collected at the 256-cell stage for analysis. (C) qRT-PCR analysis of Luc (left) or as-gfp (right) RNA levels using dsRNA precipitate. (D) Degradation dynamics of injected antisense probes. The actb2-as-P3 and as-gfp probes were injected, each at 100 pg per embryo, at the one-cell stage. About 40 embryos were collected at each desired stage. Total RNA was intramolecularly ligated before qRT-PCR detection. (E-G) qRT-PCR analysis of exogenous Luc (E) or endogenous acb2 (F,G) levels. One-cell embryos were injected with indicated materials and harvested at desired stages for analysis. Injection doses (per embryo): dSmR3nd-GFP protein, 1 ng; antisense RNA probes, 100 pg or 230 pg. (H) Normal development of wild-type embryos and those injected with 300 pg actb2-as-P3 alone or together with 1 ng dSmR3nd-mCherry (dSmR3nd-mC). Embryos were injected at the one-cell stage and imaged at the shield stage and 24 hpf. The ratio of embryos with representative morphology is indicated (bottom left). Data are mean±s.d. **P<0.01; ***P<0.001; ****P<0.0001 (Welch's t-test). ns, not significant. Scale bars: 100 µm.
|