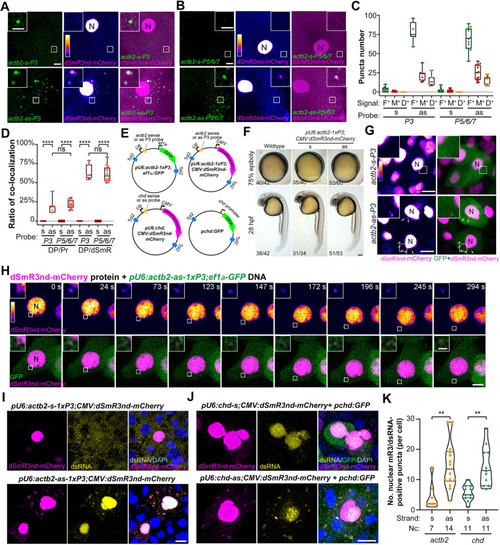
Tracking of endogenous mRNAs by dSmR3/antisense dual-color system. (A,B) In vivo tracking of maternal actb2 mRNA using dsmR3nd-mCherry together with single fluorescent actb2 probe (A) or mixed probes (B). One-cell-stage embryos were injected with 1 ng dSmR3nd-mCherry protein and 300 pg probe (for probes mix, 100 pg each) and imaged by confocal microscopy at the four-cell stage. The boxed area was enlarged in the inset. N, nucleus. (C) Total numbers of fluorescent probe puncta (F+), dSmR3 puncta (M+) and dSmR3/probe double-positive puncta (D+) in the cytosol. Data were obtained from single views as exemplified in A and B. (D) Ratios of dual-color (co-localization) labeling. DP/Pr ratio: number of dual-color puncta/number of probe-positive puncta; DP/dSmR ratio: number of dual-color puncta/number of dSmR3nd-mCherry-positive puncta. The number of embryos used for statistics was 3 (s-P3, 12 live imaging time points), 1 (as-P3, 7 time points), 8 (s-P5/6/7, 34 time points) and 2 (as-P5/6/7, 14 time points). Horizontal lines in the box plots show the median, boxes show the first to third interquartile ranges and whiskers represent the values outside the middle 50%. (E) Illustration of plasmids used for tracking of mRNAs at later stages. Tol2, Tol2 transposon LTRs. U6, ef1a and CMV represented promoters. (F) Normal morphology of embryos injected with plasmids for RNA imaging. One-cell-stage embryos were injected with plasmid pU6:actb2-1xP3;CMV:dSmR3nd-mCherry (20 pg per embryo) and observed at the indicated stage. (G,H) Tracking of actb2 mRNA at the shield stage. One-cell-stage embryos were injected with 1 ng dSmR3nd-mCherry protein and 20 pg pU6:actb2-1xP3;ef1α:GFP plasmid DNA and observed by confocal microscopy at 6 hpf. (G) Example of single embryos with multiple cells. Arrows indicated dSmR3nd-mCherry-positive puncta in the cytosol. N, nucleus. (H) Time-lapse live images of a single cell. See also Movie 5. (I-K) Tracking of actb2 (I) and chd (J) mRNAs by promoter-driven expression of RNA probes and dSmR3nd-mCherry. One-cell-stage embryos were injected with indicated plasmids, each at 20 pg per embryo, and collected at the shield stage for immunostaining with mCherry and dsRNA antibodies together with DAPI staining. Confocal images are shown (I,J) and the number of mCherry/dsRNA double positive puncta in the cytosol was calculated (K). Note that, in J, the weaker dsRNA signal in the mCherry-positive nucleus in the top panel compared with that in the bottom panel might be due to those cells in different phases of the cell cycle. Nc, number of observed cells. Violin plot outlines show the kernel probability density, long and short dashed lines illustrate the median and interquartile ranges, individually. **P<0.01; ****P<0.0001 (Welch's t-test). ns, not significant. Scale bars: 10 µm (A,B,G,I,J); 5 µm (A,B insets, H); 100 µm (F); 1 µm (H inset).
|