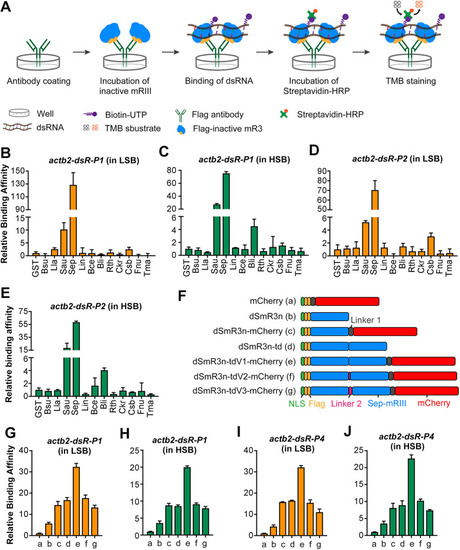
Screening of inactive mR3 proteins with high dsRNA-binding affinity. (A) Scheme of ELISA to compare relative binding affinity of inactive mR3 proteins with biotin-labeled dsRNA. (B-E) Relative binding affinity of inactive mR3 proteins with actb2-dsR-P1 (B,C) or actb2-dsR-P2 (D,E) in LSB (B,D) or HSB (C,E) buffer. mR3 origins: B. subtilis (Bsu), Lactococcus lactis (Lla), S. aureus (Sau), S. epidermidis (Sep), Listeria innocua (Lin), Bacillus cereus (Bce), Bacillus licheniformis (Bli), Ruminiclostridium thermocellum (Rth), Caldicellulosiruptor kristjanssonii (Ckr), Caldanaerobacter subterraneus (Csb), Fusobacterium nucleatum (Fnu), Thermotoga maritima (Tma). GST served as the control protein. (F) Illustration of different forms of dSmR3. Full name of dSmR3nd: dead form of Sep-mR3 with nuclear localization signal in dimer. (G-J) Relative binding affinity of different dSmR3 forms: actb2-dsR-P1 (G,H) or actb2-dsR-P4 (I,J) in LSB (G,I) or HSB (H,J) buffer. mCherry served as the control protein. The concentration of monomer protein used in ELISA is 0.8 µM, dimer protein is 0.4 µM, dsRNA is 0.02 µM. Data are mean±s.d. from three repeats.
|