Fig 1
- ID
- ZDB-FIG-200406-90
- Publication
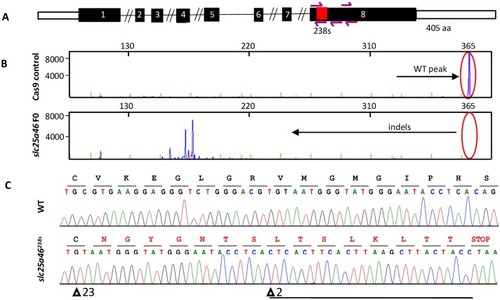
- Buglo et al., 2020 - Genetic compensation in a stable slc25a46 mutant zebrafish: A case for using F0 CRISPR mutagenesis to study phenotypes caused by inherited disease
- Other Figures
- All Figure Page
- Back to All Figure Page
|
Genome editing of |