Fig. 5
- ID
- ZDB-FIG-190802-2
- Publication
- Xue et al., 2019 - A 3D Atlas of Hematopoietic Stem and Progenitor Cell Expansion by Multi-dimensional RNA-Seq Analysis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
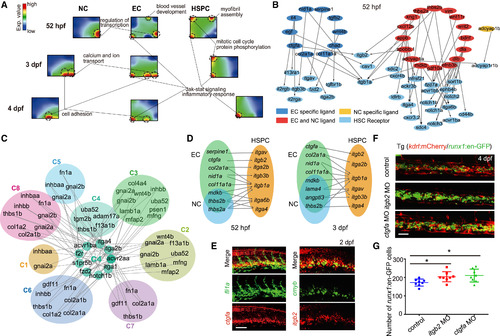
The Crosstalk between CHT Niche and HSPCs (A) SOM showing the distinct gene regulatory structure among NCs, ECs, and HSPCs at 52 hpf and 3 and 4 dpf. (B) Secretomics analysis showing the ligand-receptor regulatory relationship between HSPCs and CHT niche at 52 hpf. (C) The secretomics analysis for scRNA-seq data revealing the interaction pairs in distinct cell types, receptor-expressing C4 (proliferating HSPC) and ligand-expressing clusters. (D) The distribution of ligands and receptors in ECs (kdrl+ cells), HSPCs (CD41+cells), and NCs (kdrl-CD41- cells) at 52 hpf and 3 dpf based on bulk RNA-seqanalysis. (E) Double FISH showing the co-expression of ctgfa with fli1a, and itgb2 with cmyb at 2 dpf. Scale bar, 50 μm. (F) The confocal imaging of the transgenic line (kdrl:mCherry/runx1:en-GFP) showing the number of runx1+ cells in control, ctgfa morphants, and itgb2morphants at 4 dpf. Scale bar, 50 μm. (G) The quantification of confocal imaging displayed in (F) are shown as means ± SEM. ∗p < 0.05, n = 8. The HSPCs were counted in about the eight-somite region of the CHT in control embryos and morphants.
|
| Genes: | |
|---|---|
| Fish: | |
| Knockdown Reagents: | |
| Anatomical Terms: | |
| Stage Range: | Long-pec to Day 4 |
| Fish: | |
|---|---|
| Knockdown Reagents: | |
| Observed In: | |
| Stage: | Day 4 |