|
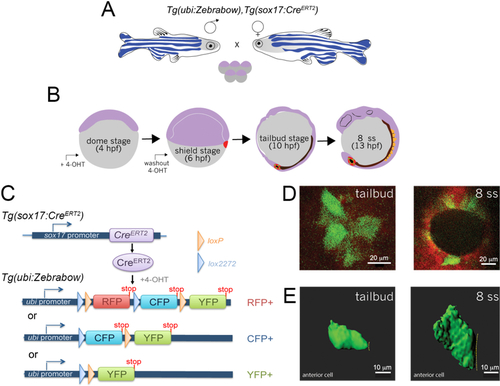
Mosaic labeling and 3D rendering of single KV cells.A. Double transgenic Tg(sox17:CreERT2); Tg(ubi:Zebrabow) zebrafish are incrossed to obtain embryos. B. Time course of mosaic labeling of KV cells. Brief treatment of double transgenic Tg(sox17:CreERT2); Tg(ubi:Zebrabow) embryos with 4-OHT from the dome stage (4 h post-fertilization) to the shield stage (6 hpf) generates low levels of Cre activity that changes expression of default RFP to expression of CFP or YFP in a subset of KV cells. C. Structure of the ubi:zebrabow and sox17:CreERT2 transgenes and the possible recombination outcomes of the Zebrabow transgene by Cre recombinase activity in KV cell lineages. Cre can mediate the deletion of sequences flanked by loxP sites (orange triangles) or variant lox2272 sites (blue triangles), leaving behind single loxP or lox2272 sites that are not cross-compatible with each other. D. Mosaic labeled YFP+ KV cells (pseudo-colored green) at the middle plane of KV at tailbud stage and 8 somite stage (8 ss). Scale bars = 20 μm. E. 3D reconstructions of single KV cells (green) using Imaris software at tailbud and 8 ss. Dashed line indicates KV lumen surface. Scale bars = 10 μm.
|