Fig. S1
|
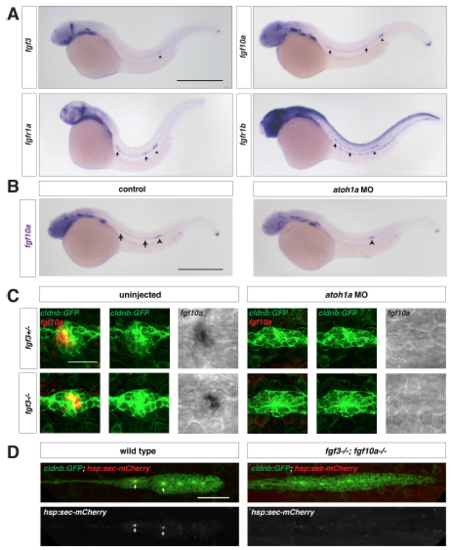
Fgf ligand expression in wild-type and atoh1a MO-injected embryos, Related to Figure 1. (A) Expression of fgf3, fgf10a, fgfr1a, and fgfr1b mRNA (purple) at 32 hpf. Arrows indicate expression in neuromasts and arrowhead denotes the position of the primordium. Scale bar = 500 μm. (B) fgf10a mRNA expression at 32 hpf in a control embryo (left) and an embryo injected with a morpholino against atoh1a (right). Arrows indicate fgf10a expression in central cells of deposited neuromasts, and arrowheads indicate fgf10a expression in the front of the primordium. Scale bar = 500 μm. (C) Maximum intensity projection of neuromast 1 in embryos of the indicated genotypes. Left, composite of GFP from cldnb:lyn2GFP (green) and fgf10a mRNA (false-colored red). Middle, GFP only. Right, fgf10a mRNA only. Scale bar = 25 μm. (D) Maximum intensity projection of the primordium in live, heat-shocked embryos of the indicated genotypes. Top, composite of GFP from cldnb:lyn2GFP (green) and secreted mCherry from hs:sec-mCherry (red). Bottom, mCherry only. Scale bar = 50 μm. |
| Genes: | |
|---|---|
| Fish: | |
| Knockdown Reagent: | |
| Anatomical Terms: | |
| Stage: | Prim-15 |
| Fish: | |
|---|---|
| Knockdown Reagent: | |
| Observed In: | |
| Stage: | Prim-15 |
Reprinted from Developmental Cell, 46(6), Wang, J., Yin, Y., Lau, S., Sankaran, J., Rothenberg, E., Wohland, T., Meier-Schellersheim, M., Knaut, H., Anosmin1 Shuttles Fgf to Facilitate Its Diffusion, Increase Its Local Concentration, and Induce Sensory Organs, 751-766.e12, Copyright (2018) with permission from Elsevier. Full text @ Dev. Cell