Fig. 7
- ID
- ZDB-FIG-120720-79
- Publication
- Clark et al., 2012 - Loss of Llgl1 in retinal neuroepithelia reveals links between apical domain size, Notch activity and neurogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
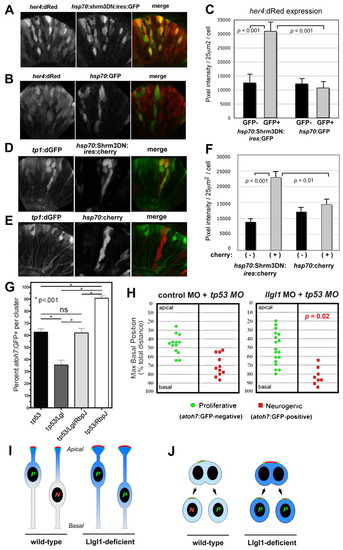
Augmented Notch signaling is required for reduced neurogenesis with loss of Llgl1. (A,B) Retinal expression of her4:dRed in 36 hpf embryos injected with plasmid DNA for hsp70:Shrm3DN:ires:GFP (A) or hsp70:GFP (B). (C) Pixel intensity measurements comparing her4:dRed expression in hsp70:Shrm3DN:ires:GFP-positive (n=58) and -negative (n=162) or hsp70:GFP-positive (n=72) or -negative (n=138) cells. (D,E) Retinal expression of tp1:d2GFP in 36 hpf embryos injected with plasmid DNA for (D) hsp70:Shrm3DN:ires:mCherry or (E) hsp70:mCherry. (F) Pixel intensity measurements comparing tp1:d2GFP expression in hsp70:Shrm3DN:ires:GFP-positive (n=73) and -negative (n=73) or hsp70:GFP-positive (n=66) or -negative (n=70) cells. Error bars in C and F represent s.e.m.; n equals total number of cells quantified from more than three embryos for each condition; P, Student’s t-test. (G) Percentage of cells within an atoh7:GFP-donor cluster that express GFP at 36 hpf. The morpholino condition is listed under each bar: 8 ng tp53 MO; 8 ng tp53 + 8ng llgl1 ATG MO; 8 ng tp53 + 8ng llgl1 ATG MO + 4 ng rbpj MO; 8 ng tp53 + 4 ng rbpj MO. For each condition, n=12 clusters were scored from 12 chimeras. P, one-way ANOVA following Tukey’s post-hoc test; ns, not significant. (H) Maximum basal nuclear position during interkinetic nuclear migration in retinal donor cells that expressed atoh7:GFP (green) or remained GFP negative (red) from donor embryos injected with either 8 ng control + 8ng tp53 MO (left) or 8 ng tp53 + 8ng llgl1 ATG MO (right). Maximum nuclear position is significantly different for neurogenic progenitors in control versus llgl1 morphants; P, Wilcoxon Rank Sum Test. (I) Schematic model showing neuroepithelial cells undergoing nuclear migration. Wild-type cells with basal nuclei become neurogenic progenitors. In Llgl1-deficient cells, the apical domain (red) is expanded and the apical-associated signals (blue gradient) are increased, causing cells with basal nuclei to divide as proliferative progenitors. (J) Schematic model showing neuroepithelial cells undergoing cytokinesis. Divisions that differentially partition the apical membrane drive asymmetric fates, whereas divisions that equally segregate the apical domain result in symmetric fates. Cells with larger apical domains, such as with Llgl1 deletion, may tend to partition this domain equally. |